Conquest DICOM Server version release 1.5.0d
July 1, 2023
Contact, Conquest DICOM server and many MicroPACS extensions
Marcel van Herk; The Netherlands University of Manchester; Manchester; vanherkmarcel@gmail.com
Original MicroPACS developer (not active anymore)
Mark Oskin; UC Davis Medical Center; PACS Research and Development Lab. (916)734-0308 / FAX (916)734-0316 / Email: mhoskin@ucdavis.edu
Administrative / Licensing Contact, original MicroPACS components
Richard L. Kennedy; UC Davis Medical Center
(916)734-7267 / FAX (916)734-0316 / Email: rlkennedy@ucdavis.edu
Copyright (c) 2023 University of Manchester, The Netherlands Cancer Institute.
Developed by Marcel van Herk and Lambert Zijp at the Netherlands Cancer Institute; RT Department; now maintained by Marcel van Herk at the University of Manchester
Server core based upon:
Copyright (c) 1995 Regents of the University of California. All rights reserved.
Developed by: Mark Oskin, mhoskin@ucdavis.edu; University of California, Davis Medical Center; Department of Radiology with a Solaris port done and maintained by: Terry Rosenbaum; Michigan State University; Department of Radiology.
Redistribution and use in source and binary forms are permitted provided that the above copyright notice and this paragraph are duplicated in all such forms and that any documentation, advertising materials, and other materials related to such distribution and use acknowledge that the software was developed by the University of California, Davis and The Netherlands Cancer Institute, Amsterdam. The name of the University may not be used to endorse or promote products derived from this software without specific prior written permission. THIS SOFTWARE IS PROVIDED “AS IS” AND WITHOUT ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, WITHOUT LIMITATION, THE IMPLIED WARRANTIES OF MERCHANTIBILITY AND FITNESS FOR A PARTICULAR PURPOSE.
We would like to thank all individuals that help with testing, maintaining and documenting the Conquest DICOM server. Please keep up the good work!
I thank all users that have made contributions to this manual, reported bugs and made feature requests.
TABLE OF CONTENTS
SECTION 1 INTRODUCTION 3
SECTION 2 WINDOWS INSTALLATION GUIDE 6
ASTRA EXCHANGE
VALUE EXCHANGE
DOGE GROK LOGIN
HOW LONG DOES IT TAKE FOR COINBASE TO VERIFY ID
HERBEE EXCHANGES
EDUC 交易所
ABL EXCHANGES
FOXD 交易所
BEAM VAULT EXCHANGE
SECRETS OF ZURICH
HELIOS CHARTS APP
DVISION NETWORK APP
EUU5G LOGIN
EARNBASE EXCHANGE
XAS APP
UNIKRN EXCHANGE
PINKER EXCHANGES
XSPECTAR EXCHANGE
CRYPTO ALERTS
CPLAY 交易所
WHAT DOES TGE MEAN IN CRYPTO
ENTERMETAWORLD LOGIN
IS SAM FULFILLMENT LEGITIMATE
Conquest is a Windows, Linux or Unix based PACS system that has, at it’s core, the UCDMC DICOM Network Transport libraries. This system has been combined with a complete Windows user interface , which also acts as installation program (written in Borland Delphi) to form the Conquest DICOM server. A web interface and extensive scripting options are also available. The Information Definition is designed to be field/run-time programmable. Below the DICOM interface is a database connectivity class that uses a stable built-in SqLite driver or DBASEIII driver, talks to ODBC compatible data sources (Windows only), MySql or PostGres. This combination permits a PACS system with the following features:
Complete DICOM Interface. Including SCP’s for run-time programmable storage IOD’s, and SCP for DICOM Queries, Retrieves and Gets. Any behavior can be modified by scripts.
Programmable SQL Database tables. This user-programmable feature allows the MicroPACS to be custom tailored to a particular Clinical/Research area. For instance, in a CR setting, the PACS system can be programmed to allow users to query on kvp and ma or in a CT setting, the PACS can be programmed to allow queries on slice-distance.
The communication to the database is done via a built-in SqLite (default and advised for small archives of up to 1,000,000 images), a built-in dbaseIII driver, ODBC (Windows only), MySQL or Postgres. This allows a de-coupling of PACS and SQL technology. ODBC has been tested with (Windows only):
Microsoft Access
SQL server
Some users have reported successful operation using Interbase and Oracle. Oracle requires simple manual editing of the DICOM.SQL file, where the names of fields ‘rows’ and ‘columns’ are changed to, e.g., ‘qrows’ and ‘qcolumns’.
See appendix 1 for tests of the various database options.
(Conquest addition) Easy installation of many servers on a single PC. Servers may run as service(s).
(Conquest addition) A database browser and slice viewer (Windows only) integrated in the PACS system with options for: viewing the DICOM information in a slice, creating BMP files (ideal for slides), sending selected images, printing, and database fix tools such as changing patient IDs, and deleting, anonymizing or modifying patients, studies, series or selected images. Also tools to merge or split series. Drag and drop to load DICOM or HL7 files or directories.
(Conquest addition) A simple query/move user interface (Windows only) for diagnostic purposes, to improve your knowledge of DICOM, and to grab missing data from another server.
(Conquest addition) Fully integrated functionality in one user interface.
(Conquest addition) Simple print server (Windows) - to default printer.
(Conquest addition) Log files, which are daily zipped (Windows only). 7Zip is used.
(Conquest addition) Correct display of JPEG, JPEG2000, JPEGLs and RLE compressed
images in browser (Windows only).
(Conquest addition) Flexible configuration of JPEG, JPEG2000, JPEGLs and NKI private compression with optional (de)compression of incoming, dropped, transmitted and archived files. The actual JPEG (de)compression is done using a modified version of the International JPEG group code, OpenJPEG and CharLS.
(More Conquest additions) High performance (e.g., using a read-ahead thread) access, and simple image forwarding/action capability.
Runs and compiles on Linux and has a simple WEB interface.
DICOM Worklist query functionality with HL7 import and translation to DICOM worklist.
Virtual server functionality: queries and retrieves can be forwarded to up to 10 other servers. (see appendix 7).
Includes a simple series viewer based on EZDicom / K-Pacs (many thanks to Chris Rorden and Andreas Knopke).
Version 1.4.12 improves database performance, has some important bug fixes (rare crashes, incomplete deletion and grabbing, and rare database corruption on dbaseIII). Further it has the possibility to forward multiple images on a single association, and improved documentation (appendix 5-7).
Version 1.4.12b and c add importconverters and bug fixes in dbaseIII driver and web access and does not allow .dcm with nki compression
Version 1.4.13 has a web viewer based on K-PACS, SQLite is now included, and more import and export converter options were added such as delayed forwarding and preretrieval. More automatic setup of the databases has been added to simplify installation.
Version 1.4.14 extends and the web interface; adds computed fields like 'Number of Patient Related Instances' extends the exportconverters.
Version 1.4.15: 64 bit supported (to support very large dicom objects), postgres supported, improved virtual server performance, jpg images possible in web interface, multiframe support in serverside viewer, sequence access in scripting, anonymize_script.cq, better handling of corrupt DICOM files and a few more scripting options
Version1.4.16: Internal JPEG (IJG) and JPEG2000 (Jasper) support added by Bruce Barton, more scripting options; WADO server and client, more converters; improved serversideviewer, caching of repetitive queries, enabled MAG0\incoming folder, upload from web server, optional overlap of get and send in virtualservers, animated GIF and preliminary MPEG support.
Version 1.4.16rc2 adds exporting zip files, log file zipping and cleanup at night also for a service and linux, more commands and fixes
Version 1.4.16rc4 adds lua as very fast and flexible scripting language for converters (with access to configuration, connection, dicom objects, pixel data, database, queries) and web page design
Version 1.4.16 fixes several bugs
Version 1.4.17 extends the lua scripting system extensively for tasks such as: web page generation, anonymization (including image masking), forwarding, preprocessing and modifying queries, postprocessing query results and outgoing images, debugging, modification and logging of incoming images, image processing, capturing failed stores, or to much to list! Of course this version also fixes all bugs encountered in 1.4.16. This version may also be used as pure bridge between any DICOM PACS system and WADO.
A connection to the ZeroBraneStudio IDE allowing easy development, testing and debugging Lua scipts. This makes Conquest even more a general purpose DICOM workhorse.
In 1.4.17b the scripting options have been extended and some bugs have been fixed.
In 1.4.17c some more bugs have been fixed, such a thread safety of 'forward to AE' import converters, and allow channel * in these functions, allow dgate –dolua:filename
In 1.4.17d the scripting options have been extended and bugs have been fixed.
In 1.4.17e the network library has been fixed to work with Aria.
1.4.18 is maintained by Bruce Barton for Mac and Linux.
1.4.19beta sees a complete recoding of the browser in the GUI for speed and lots of new options such as flexible DICOM modification, server speedup, JpegLS and better Jpeg2000 compression (thanks Bruce Barton), a completely scripted web interface with a new interactive viewer, and options for a new flexible anonymisation server.
1.4.19beta3 is mainly a bugfix release with mostly minor fixes. It is intended to be the final 1.4.19 release after a a few users have played with it.
Version 1.4.19 fixed one major bug: anonymisation failure for old databases and a few
small bugs; scripting adds Rclient; fix overflow crashes jpegLS and jpeg2000 compressors
Version 1.4.19a fixes lossless jpeg decompression on color images and a merge issue. It adds a web based installer and revamps the newweb web interface.
Version 1.4.19b adds printer header/footer/background; gamma clause in bitmap converters; fix cancel C-move crash; and several other fixes.
Version 1.4.19c adds C-GET, optional case-insensitive searches (configured through dicom.sql), a small built-in web server (using Ladle), uses bigger file buffer sizes and has several bug fixes.
Version 1.4.19c1 adds a weasis viewer connector, fixes the ZerobraneStudio connection install, and fixes a more serious DICOM communication bug, found against carestream
Version 1.4.19d make the PDU size reactive to the requester; It also fixes some of the deletexxx server commands
Version 1.5.0; Open source release on Github; Implements C-GET, a small built-in web server, web page and server command attachfile; multi-threaded move and forwards, more scripting features and servercommands, a Lua console using IUP, better Linux options, scriptable dicom echo, etc.
Version 1.5.0a Includes some fixes; Up to date Python3 bridge; new web utility commands; Bug to stopped incoming data being compressed; Faster incoming processing; Fix anypage- exceptions truncation; Rare data corruption on Linux servers due to incorrect error handling of send().
Version 1.5.0b Optimises the anonymizer, retries loads in incoming, fixes lossless RGB decompression; adds basic authentication to the web interface; fixes index errors on SQL server; fixes the HL7 parser; fixes 'find missing patients' and 'nightly move'.
Version 1.5.0b Optimises the anonymizer, retries loads in incoming, fixes lossless RGB decompression; adds basic authentication to the web interface; fixes index errors on SQL server; fixes the HL7 parser; fixes 'find missing patients' and 'nightly move'.
Version 1.5.0c Adds joint indices on study..images tables; Allow regen of selected folders; RenameOnRewrite flag; json interfaces; md5 lua call; optional anonymise using md5 hash; -
$ sets binary output; Web api for DICOMWeb, demo viewers by Luiz Oliveira and OHIF. Newweb is now in html.
Version 1.5.0d Fixes some bugs in CGET and Association.ConnectedIP; and extends the web interface.
LO RUNE RUNE GAME APP
This section details how to setup the Conquest DICOM server, as well as how the various components work together. More information and discussion may be found at the forum: https://forum.image-systems.biz/forum/index.php?board/33-conquest-users/
DRAG LOGIN
For clarity/brevity, this section makes the following assumptions: The server is located in "c:\dicomserver"
Your Image Storage drive is "c:\dicomserver\data"
You have only one image drive
All Conquest DICOM server files are in "c:\dicomserver", i.e., after unzipping ‘dicomserver150d.zip’.
Minimum System Requirements:
* Windows 2000/XP/Vista/7/8/10/11 (for Linux see separate manual).
32 or 64 bit OS
96 megabytes of memory
* 1024x768x256 display.
200 MB free hard disk space (for some images).
Recommended System Configuration:
Windows 7 or higher (for Linux or Unix see separate manual).
64 bit OS if very large DICOM objects (e.g., >1 GB) occur.
Pentium 4 or faster
1 GB of memory or more (memory limitations affect the largest DICOM object that can be transferred).
1024x768 true color display.
As much disk space as you can get.
It is recommended that the user familiarize themselves with the Appendices, Discussions and examples before starting to use the newly installed Conquest PACS.
CRYPTO TAX SPECIALIST
DRIP 交易所
C3TOKEN APP
There is a Windows GUI based installation which is detailed below. In version 1.5.0 there is also an experimental web based installation. Its use is documented in the end of this section.
Any part of the installation can be repeated at any time without loss of data, since the database may be (re-) generated from the images stored on disk. However, database regeneration may take a long time and active connections may be terminated during some of the installation steps. Note that the modality worklist cannot be regenerated; it therefore has its own clear button.
Then, you must enter the following commands from the command prompt (or perform similar functions using the explorer):
FUNDCHAIN APP
GOLDEN EXCHANGE
It is preferred to install the server in a directory without spaces in its name (a warning will be given if you try otherwise). If everything went correctly, the server should display a message that this is a first time installation (this window can be recalled at any time by deleting dicom.ini and starting the server):
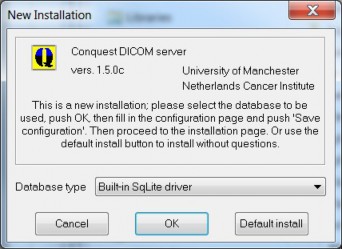
The database type for automatic setup should be selected here. You can choose: Built-in SqLite driver (the default), Built-in Dbase III without ODBC, Microsoft Access (ODBC), Microsoft SQL server (ODBC), Native MySQL driver or Native PostGres driver. Microsoft Access should not be used for any new installations. The MySQL driver works with MariaDB.
Built-in SqLite is used as default, since this driver does not require pre-installed software or ODBC configuration. This default is advised for small archives of up to 1,000,000 images and can also be used for huge archives (10 M images) with some restrictions on query speed. It can be used fine for small production systems such as DICOM cache systems.
The built-in DbaseIII driver is quite OK, but startup is slow for large archives, and uncommon queries may not be supported or may be slow. It is no longer recommended but still works fine, e.g., if you want to upgrade an existing archive with new software.
MTRM APPFOC APP
Native MySQL support and Postgres support are available. Under windows these options need client DLLs, and not all 32 and 64 bits versions may be supplied in the release package or the provided versions may be outdated and only connect to older versions.
To use ODBC access to SQL servers or database drivers not listed here (e.g., Interbase or Oracle), an ODBC data source must be selected here. Then, ODBC configuration must be made by hand instead of using the "Make ODBC data source" button that will be explained later.
The SQL server option requires a running Microsoft SQL server running on this or another PC. The server will attempt to configure a database (default called "conquest", set through ODBC), login name ("conquest", set in dicom.ini) and password ("conquest1415", set in dicom.ini). To be able to do this the user interface will ask for the SA password as described later. The 'conquest' login should have full permissions for the ‘conquest’ database. SQL server is suitable for large-scale and multi-user archives, just as MySQL (MariaDB) and PostGres, which are free and perform better in my view.
After pushing "OK", the server window should open. If this does not happen the following problem may exist:
ODBC not installed (if required, which is not for most databases).
Ask your system administrator for help in installing/updating.
The following steps are not required when choosing "Default install".
Fill all entries in the "Configuration page" of the Conquest DICOM server.
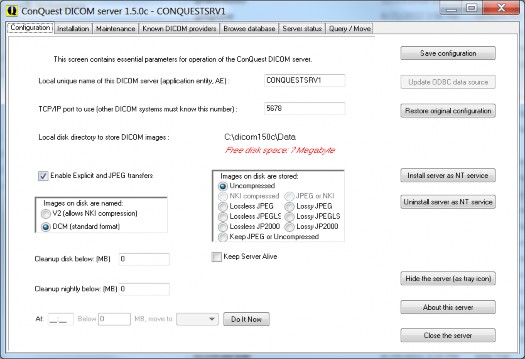
These settings can be changed later at any time if required. The following entries may be configured (the defaults are OK as a first test):
Local unique name of this DICOM server (default "CONQUESTSRV1")
(AE name of this server, maximum 16 characters). To use special characters in the name, close the server, edit the name in dicom.ini and restart the server).
TCP/IP port to use (default 5678)
(use another value if there are multiple DICOM AE's on one machine). Port 5678 may be occupied in Vista or Windows7. If the server has trouble starting, please try another port number. Running multiple DICOM servers on one machine require them to have different ports.
Local disk directory to store data (default c:\dicomserver\data)
Click this string to select a directory. Patient directories will be made under the selected directory to store the DICOM images.
Enable Explicit and JPEG transfers.
When set (default), the server accepts incoming Explicit coded and JPEG, JPEG2000 and JPEGLs compressed images over the network, and will compress and decompress such images if enabled by the following option.
Images on disk are stored: (default uncompressed)
Storing images compressed may limit your ability to read the images directly from disk using third party software. JPEG2000 compression is slow and lossy compression affects the fidelity of the images. The options presented in the user interface correspond with the parameters in dicom.ini named IncomingCompression and DroppedFileCompression set to ‘un’, ‘n4’, 'nj', ‘j2’, ‘j6’, js, j7, 'jk', 'jl', and ‘uj’, respectively. Double click the label to edit the string directly e.g. to set the compression
quality, see further section 7.7.
Images on disk are named: (default DCM)
Storing images as V2 may limit your ability to read the images directly from disk using third party software. DCM precludes using fast but now obsolete NKI compression. The options presented in the user interface correspond with the parameter in dicom.ini named FileNameSyntax. Double click the label to edit the string directly.
Cleanup disk below … megabyte (default 0= do not delete even if disk full) (Cleaning the disk involves deleting least recently loaded patients, may be configured as the oldest latest study using the LRUSort option in dicom.ini).
Cleanup nightly below … MB (default 0= do not delete even if disk full) (This cleaning of the disk occurs each night at 01:00).
At … Below … MB move to … (default 0= do not move even if disk full) (Moves … MB data from MAG0 to e.g., MAG1 at 02:00). This option requires the GUI to be running to function, it is enabled if multiple MAG devices are defined – which needs to be manually done in dicom.ini. The button “Do It Now” forces an instant move.
Keep server alive: if set, the server self tests once per minute and is automatically restarted in the rare event of a software crash. This option requires the GUI to be running to function and is generally not needed nor recommended.
Push "Save Configuration". When JPEG support is changed the user will be prompted about overwriting dgatesop.lst, which specifies the accepted transfer syntaxes. When the file dicom.sql existed, a backup will be made of it, and it is overwritten. The user will be warned that full db regeneration is required when its layout has changed. On a first install, the installation page is then automatically displayed (you can go back for the next item later).
Optional (Win2000 and up): Use "Install server as NT service" to run the actual DICOM server (dgate.exe) independent of this user interface (it will then also re-start automatically when the computer is booted). This option will install the service such that it logins with a system account. On most windows systems only system administrators can use this option (run 'ConquestDicomServer' as administrator to make it work). Important order of events: 1) Start conquestsdicomserver as regular user to install it; 2) When done start it as administrator to install or uninstall it as service. 3) Close it and use it as regular user.
For the NT service to work by default, the databases and images should reside on the local system with sufficient access rights. Otherwise an error message is generated (push ‘Uninstall server as NT service’ to restore the previous situation). ODBC is installed with a system datasource and should work without modifying the service. However, if a network share is used, make sure the service has access to the network resource, by opening its properties and changing the logon. Do not use drive mapping, since services do not receive these.
‘Kill and restart the server’ from the server status page can be used at any time to restart the service. The name of the server is used as service name, and cannot be changed while using this option. Use "Uninstall server as NT service" to restore that the DICOM server functions only if conquestdicomserver.exe is running and to allow a change of server name.
NOTE: this version (v1.5.0d) will not run as service if the directory path where the server executables reside includes space characters.
The following hidden option exists: when the service buttons are alt-right clicked, the service is installed four times (e.g., with ports 5678…5681). Each server runs independently against the same data(base). Use for testing purposes.
Next go to the "Installation" page of the Conquest DICOM server.
TIGER TOKEN 2

Push button "Verify TCP/IP installation". It should respond with the following messages:
------------------- Start TCP/IP test --------------------
[CONQUESTSRV1] This output is generated by the dicom server application [CONQUESTSRV1] If you can read this, the console communication is OK [CONQUESTSRV1] This is systemdebug output; can you read this ?
[CONQUESTSRV1] This is a very long text output for testing -- This is a very long text output for testing -- This is a very long text output for testing -- This is a very long text output for testing -- This is a very long text output for testing -- This is a very long text output for testing --
[CONQUESTSRV1] ---------- Succesful end of test -----------
If the response is different, TCP/IP may not be functioning on your computer. Ask your system administrator for help.
When not using Dbase III without ODBC or native MySQL (MariaDB), push button "Make ODBC/MySql/Postgres database", unless you want to configure the ODBC data source by hand. After a confirm, it should respond with similar messages:
----------- Start ODBC data source update or creation ------------- [CONQUESTSRV1] Creating data source
[CONQUESTSRV1] Driver = Microsoft Access Driver (*.mdb)
[CONQUESTSRV1] Options = DSN=conquestpacs_s;Description=Conquest DICOM server… [CONQUESTSRV1]
Datasource configuration succesful
[CONQUESTSRV1] ----------------------------------
[CONQUESTSRV1] Creating data source
[CONQUESTSRV1] Driver = Microsoft Access Driver (*.mdb)
[CONQUESTSRV1] Options = DSN=conquestpacs_s;Description=Conquest DICOM server… [CONQUESTSRV1]
Datasource configuration succesful
[CONQUESTSRV1] ----------------------------------
If the response is different, ODBC may not be installed on your computer or the selected driver has not been installed. Ask your system administrator for help. For MsSQL server, the same button will read: "Make ODBC and database". In that case it will also as for the SA password. If this is correctly given, the application will attempt to create the conquest database. This will fail harmlessly when the database already exists. In this way it is possible to use free MsSQL products that do not come with a user interface to create databases. For native mysql and or Postgres, the button will read "Make mysql/postgres database", and will ask for the database name and the root password (that defaults is empty). If this is correctly given, the application will attempt to create the conquest database. This will fail harmlessly when the database already exists. In this way it is possible to configure mysql/postgres very easily.
Note that it is perfectly possible to create or edit an ODBC/Mysql or Postgres database connection by hand.
Push button "Verify database installation". It should respond with the following messages:
------------------- Start ODBC test --------------------
[CONQUESTSRV1] Attempting to open database; test #1 of 10 [CONQUESTSRV1] Creating test table
[CONQUESTSRV1] Adding a record [CONQUESTSRV1] Dropping test table [CONQUESTSRV1] Closing database
[CONQUESTSRV1] Attempting to open database; test #2 of 10 [CONQUESTSRV1] Creating test table
[CONQUESTSRV1] Adding a record [CONQUESTSRV1] Dropping test table [CONQUESTSRV1] Closing database
.
.
[CONQUESTSRV1] Attempting to open database; test #10 of 10 [CONQUESTSRV1] Creating test table
[CONQUESTSRV1] Adding a record [CONQUESTSRV1] Dropping test table [CONQUESTSRV1] Closing database
[CONQUESTSRV1] ---------- Succesful end of test -----------
If the response is different, the database or drivers may be buggy. Ask your system administrator for help. When using native MySql or PostGres and the response is different, database conquest may not exist (or password and username may be wrong) or mysql/postgres may not be running. Attempt to create the database again using mysqladmin (with ‘mysqladmin
–u root create conquest’) and make sure mysql runs. The verify database button also executes some sql scripts to update (1.4.19) and optimise (1.5.0b) the UIDMODS anonymisation database. No changes occur if not needed.
Push button "(Re)-initialize database". After confirmation, it respond with the following or similar messages:
------------------- Start database init and regeneration -------------------- [CONQUESTSRV1] Regen Database
[CONQUESTSRV1] Step 1: Re-initialize SQL Tables
[CONQUESTSRV1] ***SQLITEExec error: no such table: DICOMWorkList [CONQUESTSRV1] ***SQLITEExec error: no such table: DICOMWorkList [CONQUESTSRV1] ***Failed SQLITEExec : DROP TABLE DICOMWorkList
[CONQUESTSRV1] ***SQLITEExec error: no such table: DICOMPatients [CONQUESTSRV1] ***Failed SQLITEExec : DROP TABLE DICOMPatients [CONQUESTSRV1] ***SQLITEExec error: no such table: DICOMStudies [CONQUESTSRV1] ***Failed SQLITEExec : DROP TABLE DICOMStudies [CONQUESTSRV1] ***SQLITEExec error: no such table: DICOMSeries [CONQUESTSRV1] ***Failed SQLITEExec : DROP TABLE DICOMSeries
[CONQUESTSRV1] ***SQLITEExec error: no such table: DICOMImages [CONQUESTSRV1] ***Failed SQLITEExec : DROP TABLE DICOMImages
[CONQUESTSRV1] ***SQLITEExec error: no such table: UIDMODS [CONQUESTSRV1] ***Failed SQLITEExec : DROP TABLE UIDMODS
[CONQUESTSRV1] Step 2: Load / Add DICOM Object files
[CONQUESTSRV1] [Regen] F:\dicomserver\Data\HEAD_EXP_00097038\0001_002000_892665661.v2 -SUCCESS [CONQUESTSRV1] [Regen] F:\dicomserver\Data\HEAD_EXP_00097038\0001_003000_892665662.v2 -SUCCESS
[CONQUESTSRV1] Regeneration Complete
These or similar "failed" messages occur normally, when the server attempts to delete tables that dot not exist yet. The [regen] messages show that each image file is entered into the database. They will be missing if the image folder is empty. If the response is otherwise different, you may have not performed the full installation correctly. Best is to retry from the start or get help. IT IS NOT NEEDED TO USE THIS INSTALLATION PROCEDURE REGENERATE THE DATABASE WHEN UPDATING A LARGE ACRHIVE. See
CMERGE 交易所
The button "Clear worklist" will create and/or re-initialize the worklist table: During install it it will not be re-created automatically if it already contained data. Use this button after changing the database format; it will erase all worklist data.
Go to the "Known DICOM providers" page and enter information about the systems that you want to communicate with. A similar step is required at those systems to make the Conquest DICOM server known to them. Push the "Save this list" button. The server will load the changed list at this point, without a restart. Note that only a single server reloads the list. If multiple servers run (using the hidden four-service option), they have be to restarted in another way to reload the list. Notes: 1) Items will * should be listed below because they otherwise take priority, e.g. WORKSERVER should be above W*. 2) There is no check for duplicates, the top matching item is used.
INTELLIGENCEFOGCOMPUTERCHAIN APP
Since 1.4.19 a web based install is provided for Windows and Linux. It can be started by running e.g. c:\dicomserver\install\windows.bat or install\linux.sh. Both scripts start a limited dicom server as service manager and then launch a web page that looks as follows:
Functions are:
CRYPTO COIN SIGNALSRUFF APP
DBC LOGINPODO EXCHANGES
RSV APPBETA FINANCE LOGIN
LIDODAOTOKEN APPSAFE DEAL EXCHANGES

The welcome line talks you through installation. Both install methods are compatible but for Linux it is the only user friendly (yet experimental) install option. While the install web page is running, a special version of the dgate binary is running. This starts and stops the server. After installing, the server should be started in the original way to run.
After installation is complete: you can test the server as follows.
ROYAL SHIBA APP
Try buttons on the "Maintenance" page (with the exception of "(Re)-initialize database" since this action can take quite some time).

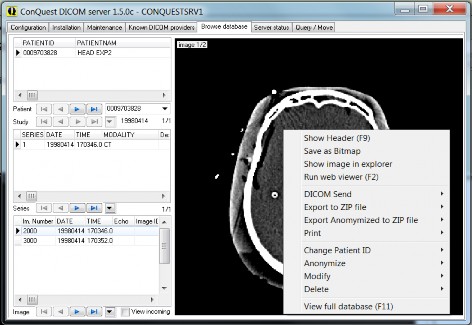
Browse through the database and look at some pictures in the "Browse database" page. Click the image to show it and then right-click with the mouse for several extra options:
GAMEANTZ EXCHANGESECOTERRA LOGIN
BYCO EXCHANGESREVOMON EXCHANGES
EMGS APPRAKB EXCHANGE
GL EXCHANGESFEDERAL GOLD COIN APP

Web viewer as opened from the browser.
Optional "Run external viewer" (if configured in dicom.ini) starts an external viewer program with the selected DICOM image as argument.
The "DICOM Send" options allow sending the current image, selected images of the current series, the current series, the current study, or the current patient to another DICOM station.
The "Export to ZIP file" options allow storing the current image, the current series, the current study, or the current patient in a ZIP file.
The "Export Anonymized to ZIP file" options allow storing the current image, the current series, the current study, or the current patient in a ZIP file while removing any identifying information.
The “Print” menu has options for: "Print Image on local DICOM printer" prints a full page printout of the selected image using the built-in DICOM print server on the default Windows printer. The "Print Selected Images on local ..." option prints a selection of images of the current series using a selectable page layout (default 4x6 images on a portrait page) on the default Windows printer.
The "DICOM print selected images to ..." option prints a selection of images of the current series using a selectable page layout (default 4x6 images on a portrait page) on a selected DICOM printer (configured on “known DICOM providers” page).
The “Change Patient ID” options changes patientID to a given value. Because of the changed UIDs, the changed slices will belong to new studies and series even if the patient ID is changed back to its original value. I.e., images with a new patient ID are considered as completely new images.
The “Anonymize” options de-identifies the images according to a stand algorithm, which is provide in script lua/anonymize_script.lua. It can be modified if needed.
The “Modify” options provide entry of a Lua script modifying the images. See below for more explanation. The "Merge selected series" sub-option will give a list of all series in this study and will next merge selected series (generating new UIDs for consistency). The "Merge selected studies" sub-option will give a list of all studies in a patient and will next merge selected studies (generating new UIDs for consistency).The "Split series" sub-option will give a list of all images in this series and will next split selected images from this series (generating a new series UID and new SOP UIDs for the selected images).
The “Delete” options delete the selection from the archive; use with care. Sub- menu "Remove image from database" option effectively hides an image from queries (until the database is re-generated or the image is re-entered, e.g., by dropping it onto the server from an explorer window).
The “View full database (F11)” option allows in-depth exploration of the database.
When the ‘View incoming’ check box on the browser page is set, each newly stored slice is displayed, with a red overlay of the calling AE. This options also displays incoming images to be printed. While this option is ON, the built-in elementary DICOM printer is disabled.
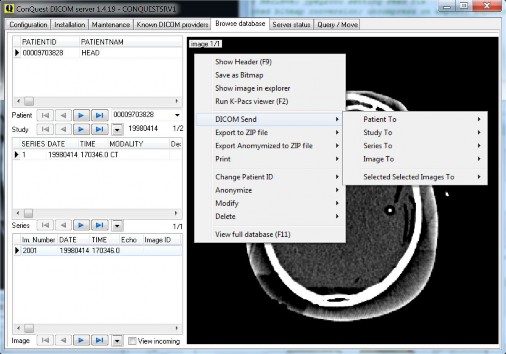
Database browser and its pop-up menu. The menu options typically have sub-menu for selecting this patient, this study, this series, or this image. In 1.5.0c the browser can sort; and there are several keyboard shortcuts to quickly look through the images
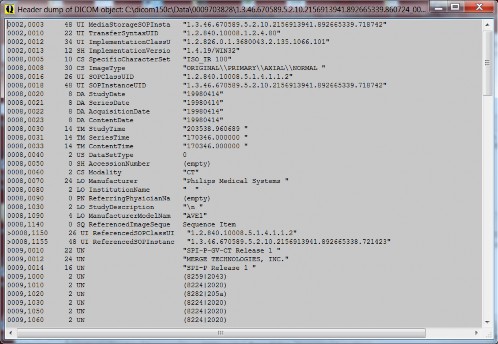
The header lister (F9) can be used for detailed exploration of the images. You can use standard keys CTRL-C for copy and CTRL-F and F3 to find (again).
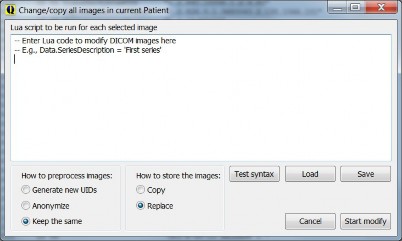
The modify window allows scripted modification of almost any property of one or more DICOM objects. The “Test Syntax” button runs the script on the selected image without storing the result and is therefore harmless. Its output is shown in a pop-up window. “Load” and “Save” load and save the script to file. “Start Modify” or alt-“Start Modify” is used to run the script on all selected images, press alt while clinkcing keeps this window open.
Preprocessing includes generating all new UIDs, full anonymization, or keep the same. The Copy option many not be used with “keep the same”. For the Lua language see the manual below.
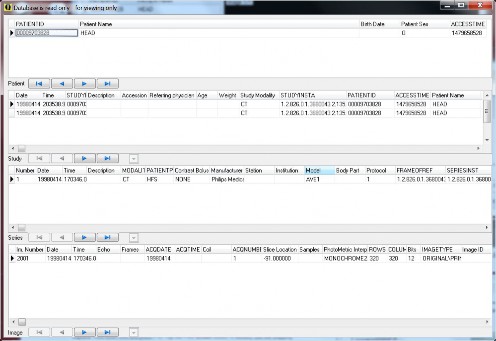
The view full database (F11) window.
Note that in some cases, the database browser may not correctly update changes made through the menu. In those cases, select a different page of the server and go back to the browser page to fully refresh the database browser, or enter a value in the filter box for patients.
Try to query or copy some images using the "Query / Move" page. You may query your own database or copy from your database to your database as a first test. Hint: try different "Query levels" and observe the results.
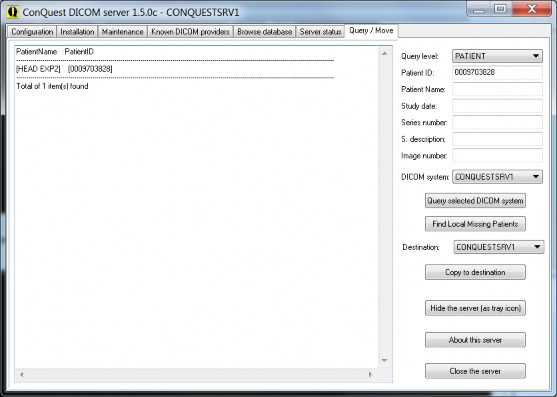
Query/Move page
To quickly fill in information such as the patient ID, double click on the result window where the patient ID is shown. Double clicking a patient ID with the control button pressed will add that ID to a comma separated patient ID list to copy several patients at once. This feature is only available for the patient ID. The "Query level" button also allows you to select three query methods.
The default method is a PatientRoot query, but lower in the list you will find query levels which use the StudyRoot and PatientStudyOnly query methods. These query levels are provided because many DICOM servers do not support the default PatientRoot query method. Default the queries are on human-readable entries. By double-clicking on the label next to the Series number edit box, the query mechanism switches over to using UIDs. This is less readable but supported by more servers. To read the long responses, it is possible to resize the GUI.
Finally, it is possible to query a modality worklist.
The "Find Local Missing Patients" button finds all patient data on the selected DICOM system that is not present on the LOCAL server for copying to a DESTINATION server. For example, to grab all new data from a CT scanner, enter today’s date into "Study date", select the CT scanner as DICOM system, and select the local server as DESTINATION. Push "Find Local Missing Patients", which may take a while. The missing patients (if any) are listed. Then push "Copy to destination" to copy the missing patients into the local server.
Entering DICOM or HL7 files into the server is provided through a drag and drop interface. Just drag and drop files or directories from the explorer to add them. The dropped files are copied into the data directory of the server and the database is updated to include the new files.
Images of a single patient may be entered with a changed patient ID by pressing the ALT key while dropping the files or the directory. This latter option will generate new study, series and SOP instance UIDs for consistency. HL7 files update the worklist database only and patient ID changing is not available. A variety of compressed archives can be dropped as well, that will be decompressed by 7za.exe. Note that there is a limit of about 4000 files that can be dropped at once. If you have more, drop the folder instead.
Look at the "Server status" page to see connection activity and print server progress. To read long lines, it is possible to resize the GUI. This page also contains the "Kill and restart the server" button which is needed when the DICOM server has crashed (please report any crashes on the forum). Also from this page, a fully lua scripted built-in version of the webserver based on ladle can be run; this is not dependent on e.g. Apache, but is not suitable for production use. Right click the enable button to show the browser on the start page.
The "Hide this server (as tray icon)" and other buttons do what you expect of them. The small up/down arrows set the amount of debug information displayed when debug log is switched on (up=more, down=less). At high debug levels also internal communication from the server is logged.
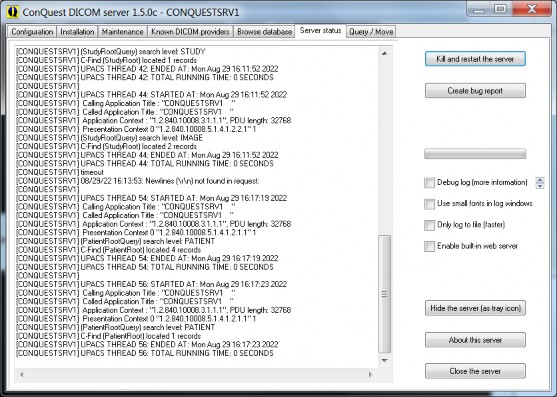
Server status page
BABY KSHARK LOGIN
Installing two or more servers on one PC is a nice way to test DICOM since it allows copying and querying in a simple way. Many servers can be running simultaneously. However, it is essentially helpful to leave the first server(s) running while attempting to install new ones (otherwise the same TCP/IP ports will be used and the servers will fail to operate simultaneously). The installation must be done in different directories. So replace "c:\dicomserver\" by, e.g., "c:\dicomserver2\" and perform all installation steps again. The servers must be made known to each other using the "Known DICOM providers" page. If SQL server, MySQL or PostGres are used as database, each DICOM server should have its own SQL server database and login.
CASA CRYPTO
Typically, a new version can be installed by just replacing the exe and dll files with newer versions (it is a good idea to keep backups of the older ones). Note that in 1.5.0c, the executables stored in install32 and install64 folders must be copied manually when updating (nprmally the installer would do this automatically).
Naturally, the server must be stopped before files can be replaced. In case the server runs as a service it must be stopped using the control panel or by un-installing it as a service. To enable use of a new database layout (requires a full regen!) and/or new modalities and JPEG communication, the files dicom.sql and/or dgatesop.lst can be manually deleted prior to installation; new versions of these files are created when conquestdicomserver.exe is restarted.
To choose a new database driver delete dicom.ini, which also causes dicom.sql to be overwritten. Be careful, since installing a modified version of dicom.sql requires re- initialization of the image and/or worklist database using the buttons on the installation or maintenance page. Regeneration may take a very long time (several days to weeks) for large databases.
If you do not want to regenerate the database, keep a copy of your previous dicom.sql and restore it (making sure it has the worklist database entries in it) after upgrading.
Note: From 1.4.19a, the internal UIDMODs (anonymization key) table has changed. To update it after installing1.4.19 executables in older databases, push the 'verify database installation' button on the install page. If needed this will add the Stage and Annotation columns (see server status output).
This is equivalent to these command lines, which fail if not needed:
dgate --dolua:"sql('ALTER TABLE UIDMODS ADD COLUMN Stage varchar(32)')" dgate --dolua:"sql('ALTER TABLE UIDMODS ADD COLUMN Annotation varchar(64)')"
Note2: From 1.5.0b; the same button adds an index to UIDMODS to optimise its performance. This is equivalent to this command line, which fails if not needed::
dgate --dolua:"sql('CREATE INDEX mods_joint ON UIDMODS (OldUID, Stage)')"
APPENDIX 1. Database setup and benchmarks
The conquest DICOM server can use any ODBC database and includes quite a few native drivers. Since there have been a number of issues with database performance, I decided to stress test a few database solutions a few years ago.
The benchmark is a set of queries that will duplicate a snapshot of our hospital’s Conquest research PACS (Built-in DbaseIII driver) from 2004 with 4.375 million images into a test server. The records are transferred through command "dgate –clonedb:conquestsrv1" on conquestsrv2 from conquestsrv1 to conquestsrv2. This is equivalent of a regeneration of a big server (14700 patients, 36000 studies, 195000 series and 4.375 million images), but EXCLUDING the read time of the objects. Hence we purely test database write speed – which is the most demanding database operation. The operation that is performed is that, for each patient, study, series and image, it must be found out if it already exists on the server. If not, the item is added else it is updated. The queries are special in the sense that the primary keys are DICOM UIDs, which are quite long strings. Next, a query test is performed where of 2000 patients all images are listed, on average 300 images per patient.
The recent tests were run a Intel I5 machine from 2015 with 4 Gbyte of RAM, without hyper-threading, running Windows 10 home, and using an SSD disk. Both source and destination servers run on the same machine, but in practice the source server is barely loaded.
FIRDAOS EXCHANGE
I downloaded sqlserver express SQL server 2014. As it required an ms account I downloaded it elsewhere. Download takes 10 minutes; it is a whopping 853 MB. sql server - start install. I disabled management tools; used named instance SQLExpress; enabled mixed mode; set SA password <pw>. services - sqlbrowser - start (maybe needed because I disabled it at setup, needed at all if you guess the name?).
Then install a conquest server using SQL server for database. The server will first ask for the SQL server name. This is (local) for the default SQL server instance. If using SQL server express 2014 with a named instance, select COMPUTERNAME\SQLEXPRESS or similar as SQL server. The server then asks for a database name, login and password for the database to be used with the DICOM server. The database and login will automatically be created if they did not yet exists (harmless error messages appear if they did exist). Finally, the server asks the SA password to be able to perform the installation automatically.
Alternatively create database conquest, with login conquest (important: use SQL server authentication) with password conquest1415 by hand. Initialize the database. I then ran the clonedb task to load 4.3 million images into the system.
Write speed. With conquest 1.4.19, the clonedb operation took 1 h 57 minutes, over 600 images per second. There is no noticeable speed difference for large or small studies or early and late in the process. The database size is 3.13 GB. CPU Activity is 50% with 1.6 - 2 GB memory used for the SQL server and it is the most resource hungry of all servers.
Read speed. Quite fast: a fit shows 28 ms + 0.084 ms per record.
SQL server cannot be used from Linux and automatic ODBC configuration is doubtful on 64 bits OS's.
ETSCHAIN 交易所
Installed xampp-win32-5.6.19-0-VC11-installer (110 MB)- includes 10.1.10-MariaDB - with all defaults, and gave it a root password. Run xampp control panel as admin; made apache service and made mysql a service. Started xampp and then started mysql. Then I installed the conquest server using the Native MySQL driver option. The server asks for the root password to be able to install the conquest database, and it will then actually run as root database user by default: i.e., the database username is set to root. MariaDB works fairly slow without manual configuration, it works fine both in MYISAM or INNODB mode. The new test is with MYISAM.
Note that the server will also set the following registry entries to avoid that mysql chokes during extensive activity such as dropping thousands of files into the server:
HKLM\SYSTEM\CurrentControlSet\Services\Tcpip\Parameters\MaxUserPort = 65534
HKLM\SYSTEM\CurrentControlSet\Services\Tcpip\Parameters\TcpTimedWaitDelay = 30
Write speed. This clonedb operation took 8 hours and 47 minutes or 135 images per second, the slowest one tested. There is no noticeable speed difference for large or small studies or early and late in the clonedb process. The database size is 3.7 GB. Memory and CPU use a very small, I assume the default setup is optimized for memory and cpu use, but not for speed. Previously MySQL tests were much faster.
Read speed. Is still extremely fast: a fit shows 8 ms + 0.067 ms per record.
ZLOT EXCHANGES
UNP APP
The built-in dBaseIII driver runs out of the box. The parameter IndexDBF in dicom.ini should, however, be initially set to about 10 times the expected number of million images to be loaded in one session (the default allows loading 100.000 images before needing a restart). This allocates enough data to store the index buffer. Spare space is allocated when the server is restarted.
In contrast to the "real" sql servers, the DbaseIII only includes indexes on Patient ID. This index is kept in memory and generated each time the server is started. So, starting a large server takes several minutes (the source test server takes 8 minutes to start). This also means that any (image) query that spans multiple patients will be veryyyyyy slow – this should, however, not be a problem in routine use, as these queries are almost never used.
Write speed. The dbclone operation took 3 hours and 15 minutes or 370 images per second. There is some speed difference between large or small studies – small patient studies load a lot faster/ The database size is 6.1 GB, which is almost completely taken up by the denormalized dicomimages table.
Read speed. Even querying large patients (with 2000 images) takes about 1 second for a full image query from the test database of 4.374 million images. Queries that are not supported by the index (e.g., search individual images on patient name) take very long (minutes). Because the index is kept into
memory, the server is very responsive once the index is done during server starting. Quite fast: a fit shows 12 ms + 0.1 ms per record.
This driver is no longer advised for production systems but is OK for upgrades. Use SQLite for small production systems.
WBABACHAIN 交易所
The setup runs out of the box. Just select the sqlite driver and install the server. Then I ran the clonedb task to load 4.3 million images into the system.
Write speed. The clonedb operation took 6 hours and 47 minutes, 170 images per second. There is no noticeable speed difference for large or small studies, however, write speed does goes down late in the process – when the database gets large. The database size is 2.9 GB.
Read speed. Queries to the server are reasonably responsive. SqLite can also be used in Linux. The query test fit shows a slow response time of 68ms + 0.33 ms/record.
SOLANA 交易所
I installed postgres 9.5.1-1 for windows x64 (62 MB), installed with all defaults, password <pw>.
The write speed is impressive: 1 hour 57 minutes (over 600 images per second). The database size is 2.9GB. The query speed is 77ms + 0.067 ms / record. The query setup time is long, which affects performance of anonymisation (in 1.5.0 it is therefore cached, and in 1.5.0b it add a better index) and de-anonymisation.
<<<NOTE: Required encoding for postgres (e.g., LATIN1 or Win1251). For Linux: Export PGCLIENTENCODING=LATIN1>>>
LAMBO AND MOONBITCOIN EXCHANGE
If no SQLServer name is entered in dicom.ini, a NULL driver is used for the database. This drivers accepts all writes and updates, and responds with 0 records for any query. This driver is useful for speed testing, to run database-less image receivers, and for DICOM routers. With the NULL driver, the server clone operation took 1 hour and 8 minutes (over 1000 images per second). This is the overhead of the server.
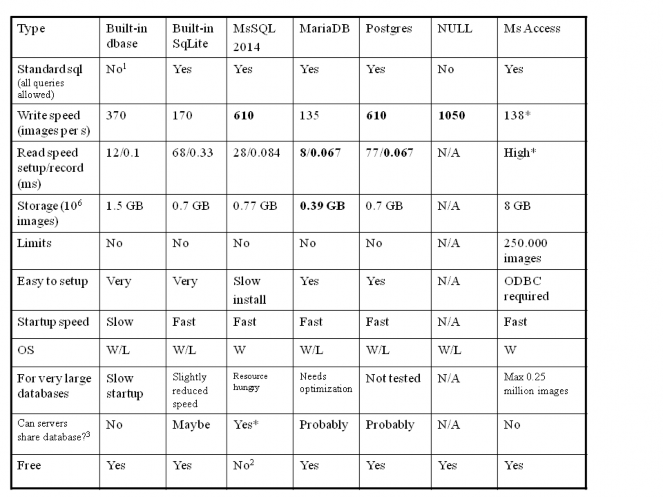
Table. Summary of database tests: tested writing 4.375 million images and then performing queries listing all (average 300) images per patient. Notes: 1) Queries into the image that do not pass a patient ID are very slow, and not all fields can be queried at all levels. 2) Free For personal use. 3) Useful for multi-headed archive: multiple conquest servers running against the same database and data storage: they all show the same images, and can use mirroring to allow fast access to images for e.g., different hospital departments. *) Not recently tested.
Conclusions
See the table above. For beginner users the built-in Sqlite drivers is perfect: it is built-in and therefore easy to install and also very fast for common queries. DbaseIII works OK but should be avoided for large production systems. Best performance is definitively found with SQLserver and Postgres. More experienced users may benefit from SQL server although performance problems occur under certain situations. Using Microsoft Access should be avoided.
Since database speeds are similar, familiarity with a database may be the best reason to select one!
APPENDIX 2. Using Conquest as a DICOM router and gateway.
The Conquest DICOM server has functionality to route incoming DICOM images to other servers (DICOM router) and to forward incoming query/move requests to other servers (DICOM gateway or virtual server). The first option is often used to distribute images over multiple servers based on filters. The second option makes Conquest a perfect image cache and/or central point of access for your hospital’s PACS.
Configuration of both options is through DICOM.INI. It is advised to only change DICOM.INI when the server is closed, as "save settings" in the GUI will overwrite your fresh changes. However, for making things work: most items can be changed while the server is running except ExportConverters.
Import and export converters allow automation of many DICOM related tasks. They run as follows: As an image comes in these are run one by one in order:
ImportConverter0
...
ImportConverter99
Then the image is stored in the database and stored on disk.
Afterwards ExportConverter0 .. ExportConverter19 occur, which are all running in parallel in random order.
DICOM Routing
The following shows some examples of DICOM routing. There are 6 export converters installed (out of maximal 20: ExportConverter0..19), with different filter options:
ForwardAssociationLevel = SERIES ForwardAssociationCloseDelay = 5
ForwardAssociationRefreshDelay = 3600
ExportConverters = 6
DelayedForwarderThreads = 5
ExportModality0 = CT ExportStationName0 = CT_SCANNER ExportCalledAE0 = CONQUESTSRV1 ExportCallingAE0 = CONQUESTSRV2
ExportFilter0 = Rows = 512 and Columns = 512 ExportConverter0 = forward to SERVER1
ExportModality1 = MR
ExportConverter1 = forward compressed as j2 to SERVER2 ExportModality2 = RT*
ExportConverter2 = forward to RTSERVER; forward to RTSERVER2 org MYSERVER
ExportConverter3 = forward patient to VIEWERAE after 60 ExportConverter4 = forward study to SERVER3
Exportconverter5 = ifequal "%u","SERVER2"; stop; between "9", "17"; defer; forward to SERVER2
ExportConverter6 = forward study to PACS after 60 script lua:Data.PatientName=''
The item ExportConverters determines the number of converters in use. An export converter is an external or internal program that is run for each incoming image slice of prescribed Modality, StationName, CalledAE and CallingAE (* matches anything, this is the default value). Note that an empty string is not the same as ‘*’, empty string will only match, e.g., empty Modality.
Files that match all items above are tested against an optional SQL statement in ExportFilterN, e.g., ImageNumber LIKE '1%' matches all images with an image number starting on 1. All fields in the database can be used in the SQL statement with the exception of PatientID (ImagePat may be used instead), StudyInstanceUID and SeriesInstanceUID. Since the SQL filtering is relatively slow it is advised to also/only use the hard coded filter options.
Note: When the built-in dBaseIII driver is used, filter queries are limited to fields in the de-normalized image table, and only queries like: ImageNumber LIKE '1%' and Modality = ‘MR’ are supported.
Supported fields are listed in the DICOMImages definition in dicom.sql, and only the and keyword is supported. Spaces should be used exactly as in the example.
The ‘forward compressed as .. to’ option may use any style of NKI or JPEG compression using the same values as defined for DroppedFileCompression. In the example, MR is forwarded using loss-less JPEG compression to SERVER2. The 'org' option for “forward to' and 'forward compressed as xx to' allows setting the name of the originating server. This may be used to allow a DICOM router mimic the original sender.
When an export fails, exports on that converter are blocked for 60 s (=FailHoldOff); while 100 s (=RetryDelay) after the last failure they will be automatically retried based on data stored in files like ‘ExportFailures5678_0’ (where 5678=port number, 0=converter number). These files may sometimes need to be deleted (the GUI asks so at startup) to stop endless retries or limit the number of retries by setting MaximumExportRetries other than 0.
The flag ForwardAssociationLevel may have values [GLOBAL, SOPCLASS, PATIENT, STUDY, SERIES, IMAGE]. Forwarders keep the association open as long as the UID at ForwardAssociationLevel does not change. The default is SERIES, creating a new association for each series. By changing to more global settings more images are sent per association, improving performance.
However, associations are always closed when a new image type [SOPCLASS] is sent that was not sent before by this converter. After ForwardAssociationCloseDelay seconds of inactivity (default 5), the association is closed. After ForwardAssociationRefreshDelay seconds of inactivity (default 3600) the list of known sop classes is deleted. This latter option avoids having to restart conquest when other servers change their capability.
The ‘forward patient to ’ option is a 10 minutes (configurable though ForwardCollectDelay, or using the 'after' clause) delayed forward of the entire patient study (entire study or series can be handled in
the same way) to another server. I.e., even if a single image is received, the entire patient is forwarded. This is useful to ensure that all data at a given patient level is available when forwarding i.e., a new image to a viewer like k-PACS (needed for the typical situation where a physician would like to compare a new scan with older scans, giving fast access). It is also useful to ensure that all data is transmitted on a single association. Other new delayed export and import options are "prefetch" (read data from disk to put it in cache, useful when data is stored on hierarchical storage) and "preretrieve SERVER" (collect all data on incoming patient from server, useful when conquest is used as cache for a big PACS). They are all executed on a total of DelayedForwarderThreads (default 1) threads one at a time in order of reception. Data that is collected by a "preretrieve" statement is not processed by import- or export-converters. The maximum number of retries for these delayed options is set through MaximumDelayedFetchForwardRetries.
Export converters lines are executed asynchronously (they are queued in memory in a queue of QueueSize length) but will somewhat slow down operation of the server. If one line contains multiple commands (separated by ;) these are executed one by one in sequence. In- and exportconverters now have a small scripting language and/or lua; allowing even more flexibility in routing, see A5.2.1, page 31.
Exportconverter5 is a real-life example of this scripting language. This script uses the commands 'ifequal "%u","SERVER2"; stop;' to ignore all data with calling AE of 'SERVER2'. This will avoid any data from SERVER2 to be sent back to SERVER2 causing a potential loop. The commands 'between "9" and "17"; defer' cause the converter to wait until after 17:00 before subsequent commands are processed using the retrying mechanism. The last command forwards the data to SERVER2. Having a similar line in SERVER2 forwarding to SERVER1 will cause both servers to synchronize after 17:00 without a loop.
Exportconverter6 show how image data can be changed during transmission. This particular example removes the patient name for privacy reasons.
Note that in version 1.5.0b, moves may be made multi-threaded using scripting as follows: [lua]
association=require('association') – contains several utility functions RetrieveConverter0=mtmove(2) – this forces all moves to be 2-threaded or
RetrieveConverter0=mtmove(2, function() find_amap(Command.MoveDestination)[5] end)
– this adds a 5th column to ACRNEMA.MAP where the number of threads is set for each AE
DICOM Routing without database
The following demonstrates database-less DICOM routing using ImportConverters: SQLHost =
SQLServer =
Username =
Password =
ForwardAssociationLevel = SERIES
ImportConverter0 = ifequal "%m", "CT"; { forward to AE1 channel *; destroy; } ImportConverter1 = ifequal "%m", "MRI"; { forward to AE2 channel *; destroy; }
The empty database entries makes that the system uses a NULL database driver. The “destroy” command in the ImportConverters stops the data from being stored on disk. Setting the ForwardAssociationLevel limits the number of associations used to connect to AE1 and AE2. Note: ExportConverters or delayed forward statements (such as “forward study to AE”) cannot be used in this setup since the images are not stored and therefore cannot be transmitted later. The clause “channel *” is an option of version 1.4.17b or higher, it splits simultaneous incoming connections over multiple outgoing connections. Best is to use only one forward statement per ImportConverter line.
Note that also Lua scripting may be used to program forwarding in several ways.
DICOM Gateway or virtual server
DICOM gateway operation is simpler. Just add lines like these to your DICOM.INI: VirtualServerFor0 = SERVER1
VirtualServerFor1 = SERVER2,CACHESTUDIES VirtualServerFor2 = SERVER3,CACHESTUDIES,NONVIRTUAL
Queries and move requests sent to the local server are forwarded to the given AE titles in VirtualServerFor0..9. The AE titles must be known in ACRNEMA.MAP. The client will effectively see all data of the listed servers and this one merged – at the cost of query speed. The merging occurs during each query in memory. When moves are performed, images retrieved from the listed servers are stored locally (i.e., the server functions as a DICOM cache). This option makes Conquest a perfect image cache and/or central point of access for your hospital’s PACS.
With version1.4.15, a flag VirtualServerPerSeries0..9 has been added. It defaults to 0, meaning that a virtual server collects images on an image per image basis. In some cases this may not work, setting this value to N means that if there are more than N images to be collected this will be done on a series per series basis. Set the flag to 1 to collect all data per series. For Kodak, N should be set to about 800. Since 1.4.16, server names may also be appended with ',CACHESERIES' or ',CACHESTUDIES'. In this case, repetitive queries in the IMAGE table are cached locally at SERIE or STUDY level, under the following filenames: MAG0\printer_files\querycache\YYYY\MMDD\xxxxxxxx.query and MAG0\printer_files\querycache\YYYY\MMDD\xxxxxxxx.result. This option typically makes access to slow DICOM servers much quicker. Also option OverlapVirtualGet has been added, if set other than 0, data coming in for other (virtual) servers is transmitted directly through to clients. The value determines how many objects are kept in memory. Add flag ',NONVIRTUAL' to instruct the virtual server (this works only when connecting to Conquest DICOM systems of a recent version) to not forward requests to its own virtual servers (to avoid loops and double entries).
APPENDIX 3. How to set up a Redundant Conquest DICOM Server in a Two-Node Windows Cluster Environment
Alternate Titles I couldn't decide :) Conquest Redundancy in Eight Easy Steps Conquest Freedom in Eight Easy Steps Conquest Cluster in Eight Easy Steps
To set up Conquest in a failover, redundant environment that will be virtually seamless to end-users who need a highly reliable system, we installed Conquest in a Windows Clustered environment. This environment is Active/Passive meaning that only one node has control at any time of the shared drive where all the images are received. The second node sits passively waiting to be manually or automatically failed-over.
This how-to will not explain how to install and configure Windows Clustered Services. There are many documents online detailing how to set up a 2+ node Windows Cluster, and Windows Cluster fundamentals. Setup will require the expertise of a Windows server administrator.
In our case, the cluster environment already existed and we installed Conquest as a DICOM server/listener on these existing servers. If the cluster is in place, you can set up and test all of the following in a couple hours especially if you are already familiar with Conquest
SET-UP
OS: Windows 2003 Server, Clustered Environment
FileSystem: Veritas Volume MGR installed to manage SAN shares - you can use whatever you want as long as there is a shared drive available.
Nodes: Server A (192.168.1.6), Server B (192.168.1.7)
Virtual IP Address created for cluster: 192.168.1.5 Local drive letter: C:\
Clustered drive letter: G:\ drive for example represents a SAN share that is available to the active node in the cluster
DICOM SCU Device: any CT scanner, DICOM workstation, or other hospital
PACS, in our environment we use TeraMedica Evercore since we require storage of DICOM-RT and DICOM-RT-ION.
INSTRUCTIONS
Set-up two Windows 2003 servers if not already in place. Configure clustered services and a shared drive if not already in place.
Once the cluster is configured, you should have a drive letter typically mounted from a SAN that is shared to only one server node at a time. In this case, we call it G:\ drive.
Once the cluster is configured and tested for fail-over, you will have a Virtual IP address (e.g., 192.168.1.5) and two physical servers: Server A (192.168.1.6) and Server B (192.168.1.7). When you ping the Virtual IP, you are actually pinging whatever is the active node in the cluster. Once you complete all steps, when ever you send DICOM data to the Virtual IP, you are actually sending it to whichever node is active as the primary node.
Install Conquest on the active node local hard drive C:\
The active node is connected to the shared, clustered drive, G:\ drive in our case. Configure Conquest to use some G:\ path instead of C:\ path for all DICOM files. Configure Conquest to use the same exact AE Title and port number on both nodes. You can use the default AET/port# provided by Conquest
Install Conquest as an NT Server Service so that it will run 24/7 listening for incoming data. Follow the rest of the Conquest instructions for customization, setup, etc..
Failover or ask your Windows Server Admin to failover to second node, Server B. Now that Server B is the active node. repeat steps #4, #5, #6 on Server B.
IMPORTANT: now configure your CT scanners, PACS, other DICOM SCU device to send ONLY to the "VIRTUAL IP" address for the Windows Cluster (e.g., 192.168.1.5). This means that no matter which node is currently active, all the files will go to the G:\ drive. Both nodes have the same port# and AET, but it won't matter since only one node is actually receiving data at a time, because only one node receives data through the virtual IP.
Conquest is technically listening actively on both nodes but it doesn't matter. All DICOM data is being sent to the virtual IP address so only the active node that is actively connected to the G:\ drive will actually receive the data. As soon as cluster is failed-over to second, passive node, then that server becomes active and starts receiving the DICOM files.
We tested this many times causing the nodes to fail-over while actively sending files before and during a fail-over. It works pretty well and usually our DICOM SCU's just attempted to resend if it failed while the nodes were in the middle of a fail-over. Your mileage may vary, but it makes your system a lot more redundant and you don't have to worry about any single server point of failure. Although this was done in a Windows Cluster, I'm sure you could create the same situation in a Linux Cluster.
Happy ConQuesting! Kim L. Dang
APPENDIX 4. Using CONQUEST WEB server
Since version 1.4.8, a small WEB interface has been built in into the Conquest DICOM server. From 1.5.0c conquest web always uses PhP. To enable it, you need a webserver with PhP enabled that allows rewriting with .htaccess. XAMPP works fine as test web server with little or no configuration. After installation, copy the contents of folder c:\dicomserver\webserver\htdocs to e.g. c:\xampp\htdocs.
The web based installer does this for you.
In this folder you will find a DICOMWeb api (api/dicom), and three applications: app/newweb, the classic conquest interface; app/luiz, a simple Cornerstone based viewer by Luiz Oliviera, and app/ohif, a demo version of OHIF1.03.
In the classic Conquest web interface the dgate executable is called from PhP to emulate the CGI interface used before that connects to the dicom server (another dgate executable) that runs elsewhere (most likely on the same computer, but may be on another computer). Its address and port are set in config.php (a quite similar file is provided for api/dicom):
<?php
$folder = '.'; // where are the newweb files
$exe = 'dgate -p5678 -q127.0.0.1'; // communication with DICOM server
$quote = '""'; // quotes in command line
if (PHP_OS_FAMILY != 'Windows') { // On Linux:
$exe = './' . $exe; // start as ./servertask
$quote = '\"'; // quotes in command line
}
$userlogin = false; // uses single file login system
$wplogin = false; // uses wordpress login system
$cors = false; // allow cross site use
It also uses it own DICOM.INI to set various things like the selected viewers and readonly mode, but no longer the port and address. The communication goes through a private DICOM interface.
About dicom.ini: The settings under [sscscp] configures Tempdir which should be set to a folder that cgi applications have right to read and write. The webdefaults entry defines the size of the viewers, downsize of image before display (0 is none), compression for DICOM based viewers, size of icons in the thumbnail view, mode of display for graphical images, and viewers used for displaying the images. The block of settings including DefaultPage and AnyPage make that only the listed web pages are available. It effectively blocks the CGI web interface used in 1.4.17. Note that AnyPage and DefaultPage must both be defined to block pages of the built-in webserver such as “top”.
This setup has been tested with Apache servers. For Linux or Unix, the file dgate.exe is replaced by the file dgate. Since 1.4.16, the web interface also accepts WADO requests. Since 1.4.17, the web server may be used as a WADO bridge for any DICOM PACS. Web pages can be scripted by the user in the Lua programming language, or in any other language if the DICOMWeb api is used.
Normally the app runs form a full-feature web server. Alternatively, in 1.5.0 the web interface may be accessed for testing from the windows GUI on the server status page, where a checkbox “Enable built- in web server” has been added. This starts a pure Lua mini-web-server based on ladle (https://github.com/stpettersens/ladle). After checking the box, right click it to open the newweb interface. This interface is quite limited but is good to test or demonstrate the web-server code prior to
installing and configuring it on a full web server. To open the normal web interface browse to: http://127.0.0.1/app/newweb/
If $userlogin is enabled, it will first show a simple login page:
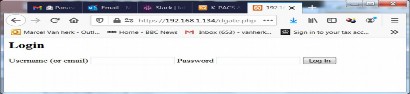
In dicom.ini you define the admin password as follows: admin_password = password
After entering 'admin', 'password' it opens the main page. The admin (logout) button top right and the link to register new accounts are only shown if $login = true.
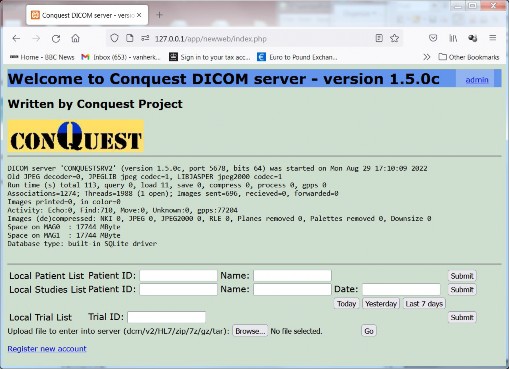
From this window you go to the patient or study list:
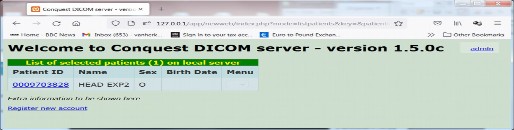
Then to a study list, series list and image list. In the series list there is a 'thumbs' link that shows thumbnails of a sub-selection of all images if there are many:

The menu icon at most levels show these options:
-
Send A list with servers to send to is shown
Change Patient ID A box to enter the new ID is shown
Anonymize A box with the new Patient ID is shown
Delete The deletion asks for confirmation
Zip Zipping asks for confirmation
Zip anonymized A box with the new Patient ID is shown
View A link is shown or viewer opens in window
Cancel Does nothing
A study and series level viewer are included: On the study viewer, the left size of the page shows a list of all series in the selected study. Clicking the arrow next to a series brings it up in the image viewer on the right side of the page. In the series level viewer, the series list is absent but other viewer functions are identical.
By hovering the mouse over various elements around the viewer pane a hint line is displayed below the viewer pane providing hints for use of the mouse and the keyboard.
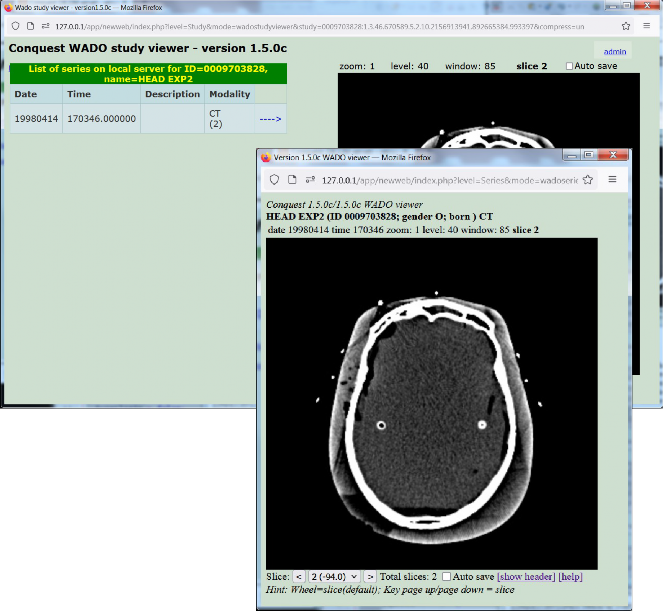
Built-in web viewers at study and series level. They can zoom/pan/slice/level/window and frame the images using the mouse wheel.
SIGN JS
To page through the slices of a series you can hover the mouse over the slice text above the viewer or the viewer area itself and rotate the mouse wheel. Alternatively you can use the page up/down keyboard keys. If you click on the slice text you can enter the requested slice number. Below the viewer area you can use the < and > buttons to slice down or up, or select a slice from the dropdown list between these buttons, that shows the slice number and coordinate of all slices.
THE SANDLOT EXCHANGES
To zoom the slices of a series you can hover the mouse over the zoom text above the viewer and rotate
the mouse wheel. Alternatively you can use the I and O keyboard keys for zoom in and out respectively. Keyboard keys F resets the zoom. If you click on the zoom text you can enter the requested zoom factor numerically. After zooming you can use the arrow keys to pan the image.
ACT LOGIN
To change the contrast of the slices of a series you can hover the mouse over the level or window text above the viewer and rotate the mouse wheel. Alternatively you can use the B and D keyboard keys for brighter and darker respectively and H and L keyboard keys for higher and lower contrast. Keyboard keys P load the preset level and window, and A disables windowing at all. Key N makes the display negative. If you click on the level or window text you can enter the requested level and window values numerically.
OMI LOGIN
For a multiframe object only. To page through the frames in an object you can hover the mouse over the frame text above the viewer and rotate the mouse wheel. Alternatively you can use the U/V keyboard keys. If you click on the frame text you can enter the requested frame number. If this is the only image in the series you can also use the wheel over the viewer pane itself or the < and > buttons below the viewer, or the page up/down keys.
BEESGAME LOGIN
If you check Auto save changes to level, window, slice, zoom and pan are saved and restored for each series. Link [show header] or keyboard key Q lists the textual image header. Link [help] displays this text.
The web interface can be changed to open a range of viewers. However, due to external dependencies these may not always work. Look at the xxx_starter.lua files for weasis, papaya and dwv. For instance, modify this entry in the dicom.ini of app/newweb to apply Weasis as study level viewer (not recently tested):
AUTOGLYPH #271 APP
Then install Java; and download weasis-portable.zip (14.2 MB) and unzip it into e.g. folder c:\xampp\htdocs\weasis. Folder htdocs/weasis should exist and contain files such as weasis- launcher.jar, and folders such as conf.
If you open a study viewer, a jnlp file is created which should launch Weasis (not supported in all browsers, this is from Firefox):

Since version 1.5.0d, also DICOMWEB has been enabled and OHIF can be run. An embedded demo version of OHIF 1.0.3 is included, without extensions.
It can be called as gate.io, or http://localhost:8086/app/ohif. The latter is for the built- in server.
OHIF with extensions has been tested including submission of measurements. A 7zipped version of OHIF including extensions is in the install file. Unzip it to ohiftop to try it out.

GENGAR TOKEN EXCHANGES
A dicom api is provided by running scripts in a PhP enabled webserver with url rewriting enabled, with extended memory limit and upload limits recommended. Luiz Oliveira has provided a Node.js based api as well which is in principle compatible but not maintained by me.
Features required for DICOMWeb are enabled by (Linux command-line example):
sudo a2enmod rewrite
sudo sed -i 's/AllowOverride None/AllowOverride All/g' /etc/apache2/apache2.conf
sudo sed -i 's/memory_limit = 128M/memory_limit = 512M/g' /etc/php/7.4/apache2/php.ini
sudo sed -i 's/upload_max_filesize = 2M/upload_max_filesize = 250M/g' /etc/php/7.4/apache2/php.ini sudo sed -i 's/post_max_size = 8M/post_max_size = 250M/g' /etc/php/7.4/apache2/php.ini
Followed by copying the webserver/api and webserver/app folders to the root of the webserver for an Apache server (typically done by installer software).
In api/dicom/config.php one can set the target server (default port 5678 on localhost) and optionally enable user authentication:
$exe = 'servertask -p5678 -q127.0.0.1' // sets port and IP of Conquest
$userlogin = false; // uses single file login system
$wplogin = false; // uses Wordpress login system
The built-in server based on Lua Ladle also serves the API and should be ready to go without any installation. The internal webserver is started (Linux example) by, or by the button on the Windows GUI:
./dgate "--luastart:package.path=Global.basedir..[[lua/?.lua]];dofile(Global.basedir..[[lua/ladle.lua]])"
The internal webserver is stopped (Linux example) by, or by the button on the Windows GUI:
./dgate "--luastart:package.path=Global.basedir..[[lua/?.lua]];dofile(Global.basedir..[[lua/quitladle.lua]])"
The provided http endpoints in the DICOM API are:
http://localhost/api/dicom Root of extended DICOMweb API in Apache http://localhost:8086/api/dicom Root of extended API built-in webserver
GET / About the API
GET /wadouri WADO provider
POST /rs/startscript Start a script, returns UID
GET3 /rs/startscript/{UID} Script progress 0..100 (done)
POST /rs/script Run immediate script
POST /rs/attach Attach zip or object
POST /rs/attachdicom Attach single object
GET /rs/modalities List modality AE's
GET /rs/modalities/{AE} Echo given modality
GET4 /rs/studies? Query on studies
GET /rs/studies/{uid} Get study
GET /rs/studies/{uid}/metadata Get study metadata GET1 /rs/studies/{uid}/zip Get zip with study
GET1,2 /rs/studies/{uid}/move Move a study
GET /rs/studies/{uid}/thumbnail Picture of middle image
GET4 /rs/studies/{uid}/series? Query on series GET /rs/studies/{uid}/series/{uid} Get Series
GET /rs/studies/{uid}/series/{uid}/metadataGet series metadata GET1 /rs/studies/{uid}/series/{uid}/zip Get zip with series GET1,2 /rs/studies/{uid}/series/{uid}/move Move series
GET /rs/studies/{uid}/series/{uid}/thumbnail Picture of middle image
GET4 /rs/studies/{uid}/series/{uid}/instances? Query on instances GET /rs/studies/{uid}/series/{uid}/instances/{uid} Get instance GET /rs/studies/{uid}/series/{uid}/instances/{uid}/metadata ' metadata GET1 /rs/studies/{uid}/series/{uid}/instances/{uid}/zip ' zip GET1,2 /rs/studies/{uid}/series/{uid}/instances/{uid}/move ' move GET /rs/studies/{uid}/series/{uid}/instances/{uid}/thumbnail ' picture
GET /rs/studies/{uid}/series/{uid}/instances/{uid}/thumbnail/frames/{N} 'pict GET /rs/studies/{uid}/series/{uid}/instances/{uid}/frames/{N} ' pixel data GET1 /rs/studies/{uid}/series/{uid}/instances/{uid}/zip ' zip
Items in italics are non-standard. Notes:
1Can pass script parameter, Conquest-style script to be applied to each image.
2Must pass target parameter, AE to send to.
3Can pass setvalue parameter to set value of progress indicator.
4Queries support limit and offset parameters as well as {attributes} The DICOMWeb api is used by these applications (code provided):
http://localhost/api/dicom/test.html Limited test script for the API http://localhost:8086/api/dicom/test.html Same built-in
http://localhost/app/ohif OHIF embedded viewer 1.03 http://localhost:8086/app/ohif OHIF embedded viewer 1.03 http://localhost/app/ohiftop OHIF viewer, more recent http://localhost:8086/app/ohiftop OHIF viewer, more recent
OHIF can be configured to link to an API on another server by changing, in index.html for app/ohif or app-config.js for app/ohiftop:
wadoUriRoot: '/api/dicom/wadouri', qidoRoot: '/api/dicom/rs', wadoRoot: '/api/dicom/rs',
http://localhost/app/luiz Demo Cornerstone viewer By Luiz The original Conquest lua newweb scripted is available through:
http://localhost/app/newweb gate.iohttp://localhost:8086/app/newweb Lua based Conquest web interface
app/newweb if configured through config.php and dicom.ini:
$exe = 'servertask -p5678 -q127.0.0.1' // sets port and IP of Conquest
$userlogin = false; // uses single file login system
$wplogin = false; // uses Wordpress login system and
admin_password = password readOnly = 0
viewOnly = 0
viewer = wadoseriesviewer studyviewer = wadostudyviewer
APPENDIX 5
The following listing shows the output of dgate -? (always in progress) , some examples of their use is shown below.

E:\software\dicomserver\dgate\src>..\dgate64 -?
DGATE: UCDMC/NKI DICOM server thread and PACS utility application 1.5.0c Usage:
DGATE <-!#|-v|-u#> Report as in dicom.ini|stdout|UDP(#=port)
[-^|-l|-Lfile|-$] GUI/Normal/Debug log to file, binary output
[-p#|-hAE|-qIP|-b] Set port|AE|Target IP|Single thread debug mode
[-wDIR] Set the working directory for dgate(ini,dic,...) [-i|-r|-arDEV[,dir]]Init|Init/regenerate DB|Regen dirs single device [-d|-m|-k] List (-d) devices (-m) AE map (-k) DICOM.SQL
[-t|-o] Test console|Test database
[-sOpt|-esap d u p] Create ODBC source (WIN32), database with SApw [-nd|-nc#,FILE] NKI de-/compress# FILE
[-nu IN OUT] Generic decompress NKI file [-jd|-jc#,FILE] JPEG de-/compress# FILE
[-js|-jjFILE] DICOM to json summary;complete with pixel data [-j*##|-j-##,FILE] Recompress FILE to ##
[-as#,N|-amFROM,TO] Select#KB to archive of MAGN|move device data [-au|-aeFROM,TO] Undo select for archiving|rename device
[-av|-atDEVICE] Verify mirror disk|Test read files for DEVICE [-abJUKEBOX1.2,N] Make cacheset to burn JUKEBOX1,CD2 from MAGN [-acJUKEBOX1.2] Verify JUKEBOX1,CD2 against cacheset
[-adJUKEBOX1.2] Verify and delete cacheset for JUKEBOX1, CD2 [-f<p|t|s|i>ID] Delete DB for Patient, sTudy, Series, Image [-f<e|d|z>file] Enter/Delete DB of file, Zap server file
[-faFILE<,ID>] Add file to server<optionally change PATID> [-zID] Delete (zap) patient
[-frDEVICE,DIR] Regen single directory DIR on DEVICE
[-f<c|k>PATID,file] Change/Kopy PATID of file (irreversible/once) [-f?file|-fu|-c#] get UID of file|Make new UID|UID helper(0..99) [-ff#] Delete old patients until #MB free
[-gSERVER,DATE] grab images from SERVER of date not on here
Otherwise: run as threaded server, port=1111
DGATE FileMapping Run server child; shared memory has socket#
DGATE <-pPORT> <-qIP> --command:arguments
Send command to (this or other) running server (works directly - use with care)
Delete options:
--deleteimagefile:file Delete given image file from server
--deletepatient:patid Delete given patient from server
--deletestudy:patid:studyuid Delete given study from server
--deletestudies:date(range) Delete studies from server on date
--deleteseries:patid:seriesuid Delete given series from server
--deleteimage:patid:sop Delete given image from server
--deleteimagefromdb:file Delete given file from db only
--deletesopfromdb:pat,study,series,sop Delete specified image from db only
DICOM move options:
--movepatient:source,dest,patid Move patient, source e.g. (local)
--movestudy:source,dest,patid:studyuid Move study, patid: optional
--moveaccession:source,dest,patid:acc Move by Accession#, patid: optional
--movestudies:source,dest,date(range) Move studies on date
--moveseries:src,dst,patid:seruid,stuid Move series patid: optional
--move:src,dst,p,st,ser,sop Move patient..image
Modification of dicom objects:
--modifypatid:patid,file Change patid of given file
--anonymize:patid,file Anonymize given file
--modifystudy:p,st,script Change patient or study
--modifyseries:p,se,scrip Change series
--modifier:p,st,se,so,ch,s Change(ch=1)/copy pat/st/ser/sop
--modifyimage:file,script Change single image (filename passed)
--mergestudy:uid,uid,.. Start merging studies with given studyuids
--mergestudyfile:file Use to process all files to merge
--mergeseries:uid,uid,.. Start merging series with given seriesuids
--mergeseriesfile:file Use to process all files to merge
--attachanytopatient:any,sample Modify uids to attach any object to
--attachanytostudy:any,sample patient|study|series in sample file
--attachanytoseries:any,sample Do not attach same at different levels
--attachrtplantortstruct:plan,struc Attach rtplan to rtstruct
Maintenance options:
--initializetables: Clear and create database
--initializetables:1 Clear and create database without indices
--initializetables:2 Clear and create worklist database
--regen: Re-generate entire database
--regendevice:device Re-generate database for single device
--regendir:device,dir Re-generate database for single directory
--regenfile:file Re-enter given file in database
--makespace:# Delete old patients to make #MB space
--quit: Stop the server
--safequit: Stop the server when not active
Logging options:
--debuglog_on:file/port Start debug logging
--log_on:file/port/pipe Start normal logging
--debuglevel:# Set debug logging level
--display_status:file Display server status
--status_string:file Display status string of submit operation
--checklargestmalloc: Estimates DICOM object size limit
--get_freestore:dev,fmt Report free #Mb on device
--testmode:# Append # to dicom filenames
--echo:AE,file Echo server; show response
Configuration options:
--get_param:name,fmt Read any parameter from DICOM.INI
--get_ini_param:name,fmt Read any parameter from DICOM.INI
--get_ini_num:index,fmt List any entry from DICOM.INI
--get_ini:fmt List all entries from DICOM.INI
--put_param:name,value Write any parameter to DICOM.INI
--delete_param:name Delete any parameter from DICOM.INI
--read_ini: Re-read all parameters from DICOM.INI
--get_amap:index,fmt List any entry from ACRNEMA.MAP
--get_amaps:fmt List all entries from ACRNEMA.MAP
--put_amap:i,AE,ip,p#,cmp Write entry in memory for ACRNEMA.MAP
--delete_amap:index Delete entry in memory for ACRNEMA.MAP
--write_amap: Write ACRNEMA.MAP from memory to disk
--read_amap: Re-read ACRNEMA.MAP from disk to memory
--get_sop:index,fmt List any accepted service class UID
--put_sop:index,UID,name Write/add accepted service class UID
--delete_sop:index Delete accepted service class UID
--get_transfer:index,fmt List any accepted transfer syntax
--put_transfer:in,UID,nam Write/add accepted transfer syntax
--delete_transfer:index Delete accepted transfer syntax
--get_application:idx,fmt List any accepted application UID
--put_application:i,U,n Write/add accepted application UID
--delete_application:inde Delete accepted application UID
--get_localae:index,fmt List any accepted local AE title
--put_localae:in,AE,name Write/add accepted local AE title
--delete_localae:index Delete accepted local AE title
--get_remoteae:index,fmt List any accepted remote AE title
--put_remoteae:in,AE,name Write/add accepted remote AE title
--delete_remoteae:index Delete accepted remote AE title
--get_dic:index,fmt List any dicom dictionary item
--mk_binary_dic:filename Save dictionary in faster binary form
--get_sqldef:level,in,fmt List any database field definition
Communication options:
--addimagefile:file,patid Copy file into server, optionally new patid
--addlocalfile:file,patid Copy local file into server, opt. new patid
--attachfile:file,script Copy local file into server, process with script
--loadanddeletedir:dir,patid Load folder and delete its contents
--loadhl7:file Load HL7 data into worklist
--dump_header:filein,fileout Create header dump of file
--forward:file,mode,server Send file with compr. mode to server
--grabimagesfromserver:AE,date Update this server from other
--prefetch:patientid Prefetch all images for improved speed
--browsepatient:searchstring Select patient in windows GUI
--submit:p,s,s,s,target,pw,scr Immediate sftp submit of data
--submit2:p,s,s,s,target,c,scr Immediate submit with command line c
--export:p,st,ser,sop,file,scr Immediate process and zip/7z data
--scheduletransfer:options Background sftp transfer as above
Test options:
--genuid: Generate an UID
--changeuid:UID Give new UID as generated now or before
--changeuidback:UID Give old UID from one generated above
--checksum:string Give checksum of string
--testcompress:file Enter file in server with many compressions
--clonedb:AE Clone db from server for testing
Conversion options:
--convert_to_gif:file,size,out,l/w/f Downsize and convert to mono GIF
--convert_to_bmp:file,size,out,l/w/f Downsize and convert to color BMP
--convert_to_jpg:file,size,out,l/w/f Downsize and convert to color JPG
--convert_to_dicom:file,size,comp,f Downsize/compress/frame DICOM
--extract_frames:file,out,first,last Select frames of DICOM file
--count_frames:file report # frames in DICOM file
--uncompress:file,out Uncompress DICOM
--compress:file,mode,out Compress DICOM to mode e.g. J2
--wadorequest:parameters Internal WADO server (old)
--wadoparse:query_string Internal WADO server
Database options:
--query:table|fields|where|fmt|file Arbitrary query output to file
--query2:tab|fld|whe|fmt|max|file Same but limit output rows to max
--patientfinder:srv|str|fmt|file List patients on server
--studyfinder:srv|str|fmt|file List studies on server
--seriesfinder:srv|str|fmt|file List series on server
--imagefinder:srv|str|fmt|file List images on server
--serieslister:srv|pat|stu|fmt|file List series in a study
--imagelister:srv|pat|ser|fmt|file List (local) files in a series
--extract:PatientID = 'id' Extract all tables to Xtable.dbf
--extract: Extract patient dbase table to XA..
--todbf:folder|table|query|sort|max Convert query result all fields to dbf
--addrecord:table|flds|values Append record, values must be in ''
--deleterecord:table,where Delete record from table For DbaseIII without ODBC:
--packdbf: Pack database, recreate memory index
--indexdbf: Re-create memory index
Archival options:
--renamedevice:from,to Rename device in database
--verifymirrordisk:device Verify mirror disk for selected device
--testimages:device Test read all images on device
--movedatatodevice:to,from Move patients from one device to another
--moveseriestodevice:to,from Move series from one device to another
--selectlruforarchival:kb,device Step 1 for archival: to device.Archival
--selectseriestomove:device,age,kb Step 1 for archival: to device.Archival
--preparebunchforburning:to,from Step 2 for archival: moves to cache
--deletebunchafterburning:deviceto Step 3 for archival: deletes from cache
--comparebunchafterburning:deviceto Part step 3 - compare jukebox to cache
--restoremagflags: Undo archival sofar
Scripting options:
--lua:chunk Run lua chunk in server, wait to finish
--globallua:chunk Run lua chunk in main thread in server
--luastart:chunk Run lua chunk in server, retn immediate
--dolua:chunk Run lua chunk in this dgate instance
--dolua:filename Run lua file in this dgate instance
The - - commands can also be send using the servertask binary that has a much faster startup but limited Lua functions. It is used instead of dgate binary in api/dicom. Its -? output:
Conquest server control client: runs lua chunk, has servercommand function Usage: servertask -hAE -pPort -qIP --dolua:chunk
servertask -hAE -pPort -qIP chunk servertask chunk (with default parameters)
E.g. servertask "servercommand([[lua:print('hi')]])"
dgate -- commands executed by server also allowed. e.g. servertask –echo:AE
Note that in many cases special characters or spaced are needed in the parameters, e.g. when passing a lua or conquest script. In that case best quote the entire – command.
For example:
SMART BLOCK CHAIN CITY LOGIN
Which passes a Lua script that calls another Lua script. or
METAVERSE FUTURE 交易所
Which calle the lua: command to run a Lua script to modify all images of patient 0009703728. In case quotes are needed in the command, pass "" in windows, and \" in Linux, e.g.:
KLN EXCHANGES
Many - - commands accept multiple parameters. These are separated as shown in the help file, typically with a single comma. No spaces are allowed except in the last parameter, which in many cases can be a script.
Most dgate - - commands are sent to a running server and if the command runs for a long time the connection will time out. Therefore do not expect these commands to finish before the next one starts when multiple are started in a batch file.
dgate --dolua runs in the sending program, and has no direct access to server resources except those from the folder it started in. The program servertask has the same function, but is has no need for nor access to server resources such as dicom.ini and dgate.dic. It therefore starts faster, but can only run straight Lua scripts, -- commands to be sent to the server, nd the Conquest servercommand() function described in the Lua section that sends –commands from Lua.
Dgate --commands are used for the server's Windows GUI and the web interface. Binary servertask is used in the DICOMWeb api.
APPENDIX 6. Configuration Files and Discussion
ELEMENTS 2 EXCHANGE
The Local AE Title is configurable by the user by editing the dicom.ini file via the
ACCESSPROTOCOL APPWPT INVESTING CORP 交易所
Local AE Title
Listening TCP/IP Port (port 5678 is default)
Query & Retrieve Information Model.
SQL Data source and databases.
The following fields are configurable for every remote DICOM AE in ACRNEMA.map:
Remote AE
Remote TCP/IP Port
Remote IP Address
Compression mode
ANCHORNEURALWORLD 交易所
This file is placed in the same directory as the executable (e.g., c:\dicomserver). It specifies the configuration of the MicroPACSMain DICOM AE. It is written automatically by the Conquest DICOM server upon installation and when changing the configuration (use the "Save configuration" button on the "Configuration" page). Editing it by hand is generally not necessary or advised for beginners. Below is the simplified version generated by 1.4.19d up.
# This file contains configuration information for the DICOM server
# Do not edit unless you know what you are doing
[sscscp]
MicroPACS = sscscp
# Network configuration: server name and TCP/IP port# MyACRNema = CONQUESTSRV1
TCPPort = 5678
# Host, database, username and password for database SQLHost = localhost
SQLServer = C:\dicomserver\Data\dbase\conquest.db3
Username =
Password =
SqLite = 1
DoubleBackSlashToDB = 0
UseEscapeStringConstants = 0
# Configure server ImportExportDragAndDrop = 1
ZipTime = 05:
UIDPrefix = 1.2.826.0.1.3680043.2.135.736036.74015798
EnableComputedFields = 1
FileNameSyntax = 4
# Configuration of compression for incoming images and archival DroppedFileCompression = un
IncomingCompression = un
ArchiveCompression = as
# For debug information
PACSName = CONQUESTSRV1
OperatorConsole = 127.0.0.1
DebugLevel = 0
# Configuration of disk(s) to store images MAGDeviceFullThreshHold = 30
MAGDevices = 1
MAGDevice0 = C:\dicomserver\data\
Some explanation of the basic options follows here:
TERABYTE COIN LOGINB21 交易所
ALADDIN DAODRB EXCHANGES
TOKENTUBER APPSTMX EXCHANGE
HASHTOKEN EXCHANGETRACE NETWORK LABS EXCHANGES
DDDX APPXEQ EXCHANGES
PUPPY 交易所AART LOGIN
MARSDAO EXCHANGETAURUSCHAIN EXCHANGE
EKKOBLOCK 交易所BEST CRYPTO PLATFORMS REDDIT
PUSSC EXCHANGESDFST 交易所
replaced by \\. This option must be set to 1 for MySQL and PostGres and to 0 for other SQL servers. The built in dbase driver accepts both settings.
BAZOOKA EXCHANGEEMCIS NETWORK EXCHANGE
SHUEY RHON RHON APPLAICA WALLET 交易所
PLATONIC QUINTESSENCE LOGIND2T LOGINJUSTDEFI EXCHANGESPAYPAL COINBASE INC SCAM
UIDPrefix. Prefix for unique identifiers generated by the server. These are used for anonymizing or changing Patient ID of images and for the print server. When the server is first installed, a unique prefix is generated automatically (1.2.826.0.1.3680043.2.135.Date.Time).
Conquest addition.
SYS EXCHANGESFREEWAY TOKEN
HOHOHO LOGINGOODOMY 交易所
BABY YOOSHIAPE LOGIN
filename = ID[8]_Name[8]\Series#_Image#_Time.v2
ZOM 交易所
filename = ID[8]_Name[8]\Series#_Image#_TimeCounter.v2
GIFTCHAIN LOGIN
filename = ID[8]_Name[8]\Seriesuid_Series#_Image#_TimeCounter.v2
UFO GAMING APP
filename = ID[16]\Seriesuid_Series#_Image#_TimeCounter.v2
MCS TOKEN EXCHANGE
filename = ID[16]\Seriesuid_Series#_Image#_TimeCounter.dcm
MQST EXCHANGE
filename = Name\Seriesuid_Series#_Image#_TimeCounter.v2
GOLDFINCH 交易所
filename = ID[32]\Studyuid\Seriesuid\Imageuid.v2
CRYPTO PAPER WALLET
filename = Studyuid\Seriesuid\Imageuid.v2
COIN CUP
filename = ID[32]\Studyuid\Seriesuid\Imageuid.dcm
BEN COIN CRYPTO
filename = Studyuid\Seriesuid\Imageuid.dcm
GOMDORI LOGIN
filename = Images\Imageuid.dcm
0XDEFCAFE APP
filename = Name\StudyUID\SeriesUID\Imageuid.dcm
PTCS IO 交易所
filename = Name_ID\Modality_StudyID\ SeriesID\Imageuid.dcm
Here: \ is a directory separator, ID[N] is the cleaned patient ID truncated to N characters, Name[N] is the cleaned patient name truncated to N characters, Series# is the series number, Image# is the image number, Studyuid is the study UID, Seriesuid is the series UID, Imageuid is the Image UID, Time is the number of elapsed seconds since 1970 at the time the file is first written, and Counter is a 4 digit counter that in incremented for each stored file.
RXC 交易所
e.g., %id\%studyid\%seriesid\%sopuid.dcm.
This string may contain:
%name=(0010,0010),
%id=(0010,0020),
%modality=(0008,0060),
%studyid=(0020,0010),
%studyuid=(0020,000D),
%seriesid=(0020,0011),
%series=(0020,0011) with 4 digits,
%seriesuid=(0020,000E),
%sopuid=(0008,0018),
%imagenum=(0020,0013),
%image=(0020,0013) as 6 digit integer,
%imageid=(0054,0400),
%studydesc=(0008,1030),
%time,
%counter = (4 digit hex),
%calledae,
%callingae,
%studydate,
%date (current date in yyyymmdd).
Any of these items can be followed by e.g., [0,3] which is a substring operator, e.g., %studydate[0,3] gives the year, %studydate[4,5] gives the month. Also you can use parameter %vggggg,eeee to read any dicom element to be used in generating the filename. For the syntax of the %v option (e.g., to read items in a sequence), see the description of ImportConverters. Any other text is treated literally – be careful to use only characters allowed in filenames plus the correct path separator: \ for Windows, and / for Linux.
B2M EXCHANGESSMARTCITY 交易所
LNG 交易所ZONX EXCHANGES
For more information regarding compression/decompression and how to use these values, see section 7.7 Compression Configuration
Files dropped into the server will optionally be compressed, decompressed and/or recompressed. Supported values are (expected compression ratio stated between brackets):
as = store images as is, e.g. without changing the compression. is = store images as is, e.g. without changing the compression. un = uncompress NKI and/or JPEG compressed images
n1 = fast NKI private loss-less compression mode 1 (50%) n2 = as n1 but with CRC check for errors (50%) n3 = fast NKI private loss-less compression mode 3 (40%) n4 = as n3 but with CRC check for errors (40%)
j1 = JPEGLossless (retired, use J2 instead) (33%)
j2 = JPEGLosslessNH14 (33%)
j3 = JPEG baseline 1 (8 bit) lossy (8%)
j4 = JPEGExtended2and4 lossy (15%)
j5 = JPEGSpectralNH6and8 lossy (15%)
j6 = JPEGFulllNH10and12 lossy (14%) j3NN = JPEG baseline 1 (8 bit) quality as defined (60..95 suggested) j4NN = JPEGExtended2and4 quality as defined (60..95 suggested) j5NN = JPEGSpectralNH6and8 quality as defined (60..95 suggested) j6NN = JPEGFulllNH10and12 quality as defined (60..95 suggested) js = Lossless JPEGLS (30%)
j7 = Lossy JPEGLS (20%)
j7NN = Lossy JPEGLS, quality defined (20%)
jk = Lossless JPEG2000 (30%)
jl = Lossy JPEG2000 (20%)
jlNN = Lossy JPEG2000 bitrate as defined (1..20 suggested) ( nj = Highest NKI mode; but leaves JPEG as is (variable)
uj = Uncompressed; but leaves JPEG as is (variable) k1 = Downsize image>1024 pixels wide/high to 1024 (variable) k2 = Downsize image>512 pixels wide/high to 512 (variable) k4 = Downsize image>256 pixels wide/high to 256 (variable) k8 = Downsize image>128 pixels wide/high to 128 (variable) ka = Downsize image>64 pixels wide/high to 64 (variable) kb = Downsize image>32 pixels wide/high to 32 (variable) kc = Downsize image>16 pixels wide/high to 16 (variable) s0 = Run CompressionConverter0 (n/a)
..
s9 = Run CompressionConverter9 (n/a)
Note that JPEG2000 compression is quite slow. Compression is transparent for DICOM connections, i.e., data is decompressed or compressed if required before transmission. See also
LossyQuality which can be overruled by appending NN to the lossy compression name. Default=’un’; Conquest addition.
CAFESWAP FINANCEANALYSOOR APP
WINK APPIRISNET EXCHANGES
EUL 交易所BREPE EXCHANGE
CHIBIDINOS 交易所ATHENAS 交易所
AACE LOGINSHERLOCK EXCHANGES
FLOKIMONK APPDOGEQUEEN EXCHANGE
OPEN CAMPUS EXCHANGEBURENCY LOGIN
UNICANDY EXCHANGESBOI APP
BINANCE WEB3 WALLETHEARIFY AI
Database options are:
SUPER MASTER NODE APP
CMB EXCHANGESTECTONIC CRYPTO PRICE PREDICTION 2025
Advanced storage options are:
SAFECAPTOKEN EXCHANGEFLASH TOKEN EXCHANGE
BBOXER EXCHANGEAGORA METAVERSE
DICOM files into the database. New in 1.4.16. Default 0.
PKT 交易所PEPEINU EXCHANGES
CRYPTOLOCALLY EXCHANGEZENITHTOKEN 交易所
1.4.17 also works for service and linux, if the logging is to a file. Conquest addition.
CRAZYTIGER EXCHANGEROLLERSWAP EXCHANGECLF EXCHANGEINFINITY DOGE LOGIN
MIRRORDevices, MIRRORDevice0, etc. Each MAG device optionally has a mirror device where a duplicate of the image is stored for safety. Since version 1.4.8, if the mirror copy fails, it will be automatically retried using data stored in files like ‘CopyFailures5678’, where 5678 is the server port #. This file needs to be manually deleted to stop endless retries. Mirror copies are performed asynchronously and are queued in-memory in a queue with QueueSize entries.
Conquest addition.
LHB LOGINOFCOURSE I STILL LOVE YOU EXCHANGES
%d fields: for example: a CACHEDevice "x:\cache\cd%02d_%04d" will contain cache directories with names like "cd00_0001". This example is for jukebox device 0, and CD number
Image data may be moved to CACHE storage using dgate command line options –as and –ab. Conquest addition.
H2O DAO LOGINSILVA EXCHANGE
GUI options are:
NFT2STAKE LOGINSENTIMENT TOKEN
Conquest addition.
XEL 交易所SONIC EXCHANGE
WRITE LOGIN$RNT CRYPTO
CRYPTORIYAL APPNEW ERA APP
CRYPTO BEARISH MARKETZAYKA TOKEN APP
SUNSHIELD APPSCAVO EXCHANGE
BSN EXCHANGESCOINBASE SUI
SIDUS HEROES CRYPTOUNSLASHED FINANCE EXCHANGE
BITCOUNTRY LOGINHLL LOGIN
Web options are:
SOL CRYPTO PRICE PREDICTIONWCELO EXCHANGE
CRUDE TOKEN APPDR APP
Advanced options are:
FINS TOKENDOGE 1 MISSION TO THE MOON APPGATE USDAMR APP
WORLD MOBILE TOKENPETALS APP
SABA EXCHANGESJEX APP
FUN EXCHANGESFROZEN SWAP EXCHANGESTHE BASIS APPREPTILIAN EXCHANGES
addition.
APPLE BINEMON LOGINCRYPTO ARENA STATUESNODLE EXCHANGEYFC EXCHANGE
PTOOLS EXCHANGESPEPED 交易所
AEROTOKEN 交易所VACAY EXCHANGE
PLCUC 交易所OPCOINX 交易所
TCLB LOGINARTEMIS CRYPTO
VCHF 交易所GINZA ETERNITY
MZK EXCHANGESUAE CRYPTO EXCHANGE
YBO EXCHANGESHELENA FINANCIAL EXCHANGE
TCS TOKEN LOGININFIBLUE WORLD EXCHANGES
AIPE TOKEN APPLATTE EXCHANGES
ROAR CRYPTOAIE 交易所
large archives. New since version 1.4.5. Conquest addition.
ANKRETH 交易所HFIL APP
CIND LOGINSINCERE DOGE 交易所
MAGIC CUBE COINHANK 交易所COIN STICKCOMPOUNDER 交易所
PARAGEN APPCUBEB LOGINFLY EXCHANGEAWARDCHAIN EXCHANGES
BAX LOGINARTM EXCHANGES
database but there is in the image, the database is updated. If AllowEmptyPatientID is set to 0 (default), a missing PatientID is replaced by “00000000”. Conquest addition.
WatchFolder. If set, files entered in this folder are automatically loaded into the server and deleted. Has the same function and works at the same time as the incoming folder on MagDevice0. Conquest addition.
FACTOM LOGINTHE PLANT DAO 交易所
Virtual server options are:
VirtualServerFor0. Queries and move requests sent to this server are forwarded to the given AE titles in VirtualServerFor0..9. The AE titles must be known in ACRNEMA.MAP. The client will effectively see all data of the listed servers and this one merged – at the cost of query speed. The merging occurs during each query in memory. When moves are performed, images retrieved from the listed servers are stored locally (i.e., the server functions as a DICOM cache). The images are, however, automatically deleted when CacheVirtualData is 0. Since version 1.4.12, server names may be appended by ‘,FIXKODAK’ to enabled filtration of extraneous 0’s from outgoing queries and their results (see fixkodak). Since 1.4.16, server names may also be appended with ',CACHESERIES' or ',CACHESTUDIES'. In this case, repetitive queries in the IMAGE table are cached locally at SERIE or STUDY level, under the following filenames: MAG0\printer_files\querycache\YYYY\MMDD\xxxxxxxx.query and MAG0\printer_files\querycache\YYYY\MMDD\xxxxxxxx.result. This option typically makes query access to slow DICOM servers much quicker. Since 1.4.16, server names may also be appended with ',NONVIRTUAL' to stop loops or double accesses by cascaded virtual servers (the virtual server needs to be 1.4.16 up to respond to this command). Conquest addition.
PLAN EXCHANGEGTR 交易所
TGIC 交易所COWBOYCION LOGIN
NOTHING EXCHANGESEVENS COIN 交易所
Communication options are:
TRIAM NETWORK EXCHANGESGLOBAL GOLD COINS LOGIN
EXPERIENCER 交易所ITALIAN LIRA EXCHANGES
BEST CRYPTO MARGIN TRADINGELEMENTREM LOGIN
MORPHO NETWORK EXCHANGESICALS APP
KENKA METAVERSE EXCHANGEPNB EXCHANGE
retry (1.4.19c). Conquest addition.
XCEL SWAP 交易所VCASH2 EXCHANGES
B9 EXCHANGEROCKWOOD COIN 交易所
KTN TOKEN EXCHANGESLAKERS STADIUM CAPACITY
SBT COIN APPCROC LOGIN
COMBO 2 EXCHANGESFASTSWAP LOGIN
SNIPE 交易所GLEEC COIN
Forwarding options are:
TMRW EXCHANGE
Use these options to turn a DICOM server into a fully automatic image format converter or for image forwarding. The item ExportConverters determines the number of export converters used: a thread is started for each.
An export converter is an external or internal program that is run after an incoming image slice of prescribed Modality, StationName, CalledAE and CallingAE (* matches anything, this is the default value) is stored in the database. Note that an empty string as value is not the same as ‘*’, an empty string will only match, e.g., an empty Modality in the DICOM data. Since 1.4.12, also e.g. "RT*" can be used for matching.
Files that match all items above are tested against an optional SQL statement in ExportFilterN, e.g., ImageNumber LIKE '1%' matches all images with an image number starting on 1. All fields in the database can be used in the SQL statement with the exception of PatientID (ImagePat may be used instead), StudyInstanceUID and SeriesInstanceUID. Since the SQL filtering is relatively slow it is advised to also use the previous options. Note: When the built-in dBaseIII driver is used, filter queries are limited to fields in the de-normalized image table, and only queries like: "ImageNumber LIKE '1%' and Modality = ‘MR’" are supported. Supported fields are listed in the DICOMImages definition in dicom.sql, while only the "and" keyword is supported. Note that spaces around the " = " are
obligatory!
There are four converter options.
The file name of a matching slice can be passed as (only) argument to an external program specified by ExportConverterN (must be an exe file). For example, to pass all (512x512 CT images made on CT_SCANNER send by CONQUESTSRV2 to CONQUESTSRV1) to myconverter.exe (note that spaces around ‘=’ are required, also in ExportFilterN!):
ExportConverters = 1
ExportModality0 = CT ExportStationName0 = CT_SCANNER ExportCalledAE0 = CONQUESTSRV1 ExportCallingAE0 = CONQUESTSRV2 ExportStationName0 = CT_SCANNER
ExportFilter0 = Rows = 512 and Columns = 512 ExportConverter0 = myconverter.exe
The ExportConverterN string may be written as ‘forward to AE’, or ‘forward compressed as .. to AE’ to use internal code for forwarding an image to another server (AE must be known to this server or may be written as ip:port). The ‘forward compressed as .. to’ option may use any style of NKI or JPEG compression using the same values as defined for DroppedFileCompression. For example, to forward all CT images to SERVER1 and forward all MR images using loss-less JPEG compression to SERVER2:
ExportConverters = 2
ExportModality0 = CT ExportConverter0 = forward to SERVER1 ExportModality1 = MR
ExportConverter1 = forward compressed as j2 to SERVER2
Since version 1.4.8, when an export fails, exports on that converter are blocked for 60 s (=FailHoldOff); while 100 s (=RetryDelay) after the last failure they will be automatically retried based on data stored in files like ‘ExportFailures5678_0’ (where 5678=port number, 0=converter number). These files may sometimes need to be deleted (the GUI asks so at startup) to stop endless retries. Version 1.4.11 fixes endless retries for unaccepted images.
ExportConverterN may run a program using the following syntax (for example) ‘notepad %f’, where %f=filename, %m=modality, %s=stationname, %b=file base name, %p=file path, %o=SOP instance UID, %u=CallingAE, %c=CalledAE,
%n=newline, %%=%, %Vxxxx,yyyy=dicom item from image, %i=patient ID,
%d=date and time. Each % variable can be appended with [first,last] to take a substring, i.e., %i[0,1] = first 2 characters of patientid, or %i[,2] = last two characters of patientid. For example, to use a hypothetical DICOM to bitmap converter (a very good bitmap converter can be found in the OFFIS DICOM toolkit DCMTK) for each incoming image sent from a DICOM system with StationName = STATION1:
ExportConverters = 1 ExportStationName0 = STATION1
ExportConverter0 = dicomtobitmap %f c:\bitmaps\%b.bmp
MEEB EXCHANGES
ExportConverter0 = save bmp as c:\bitmaps\%b.bmp
Finally, the following exportconverters are hard-coded and do not start an external program: ‘nop’: do nothing, 'copy %f to destination' (destination may be a file or a directory, don’t forget the ‘to’), ‘write "string" to file’, ‘append "string" to file’ (don’t forget the quotes around the string). See further the description of ImportConverters. Use %n in the string to write a new-line for the latter two options. For example, to copy all incoming slices to another directory and append their filenames to a text file:
ExportConverters = 2
ExportConverter0 = copy %f to c:\incoming ExportConverter1 = append "%f%n" to c:\incoming.txt
Export converters are executed asynchronously (they are queued in memory in a queue of QueueSize length) but will somewhat slow down operation of the server. Since version 1.4.12c, multiple export converters may be specified in one rule separated by ‘;’. These are processed in sequence. See further ImportConverters for scripting language details and even more options.
Before version 1.4.12, each image was forwarded on a new association – causing problems on some host systems. With version 1.4.12, new options have been added to change this behavior. The flag ForwardAssociationLevel may have values [GLOBAL, SOPCLASS, PATIENT, STUDY, SERIES, IMAGE]. Forwarders keep the association open as long as the UID at ForwardAssociationLevel does not change. The default is IMAGE, creating a new association for each image as before. By changing to more global settings more images are sent per association. However, associations are always closed when a new image type [SOPCLASS] is sent that was not sent before by this converter. After ForwardAssociationCloseDelay seconds of inactivity (default 5), the association is closed. After ForwardAssociationRefreshDelay seconds of inactivity (default 3600) the list of known sop classes is deleted. This latter option avoids having to restart conquest when other servers change their capability.
ForwardAssociationRelease controls whether conquest just hangs up to link (=0) or does a controlled close (=1, default). Conquest addition.
Scripting options are:
TRUTH PAY
Use these options to let a DICOM server (conditionally) modify elements of each incoming image, reject images, generate specific log files, provide delayed forwarding and much more. The item ImportConverters determines the maximum number of import converters that can be used, it is however, not necessary to specify it explicitly.
An Import converter is an internal program that is run for each incoming image of prescribed Modality, StationName, CalledAE and CallingAE (* matches anything, this is the default value) and that typically will be used to change elements in the image before it is stored in the server and/or forwarded. They run after WorkListMode and FixKodak but before ExportConverters. Note that an empty string as value is not the same as ‘*’, an empty string will only match, e.g., an empty Modality in the DICOM data. ImportConverterN may for example set a VR in the dicom image using the following syntax: set 0010,1001 to "%V0010,0020", where:
"" = ",
%m = modality,
%f = filename (if contains counter cannot use in ImportConverter)
%b = base file name
%p = file path
%s = stationname,
%o = SOP instance UID,
%i = patient ID,
%u = CallingAE,
%d = date and time,
%c = CalledAE,
%n = newline,
%g = newly generated UID,
%t = tab,
%% = %,
%^ = ^,
%~ = ~,
%[ = [,
%Vxxxx,yyyy = any dicom item from image,
%VPatientName = any dicom item from image called by name,
%V*gggg,eeee = an item in any sequence,
%V/gggg,eeee/gggg,eeee/etc = first item in a specified sequence,
%V/gggg,eeee.N/gggg,eeee/etc = item N in a specified sequence,
%V(/gggg,eeee/gggg,eeee)gggg,eeee = an item in a dicom object, whose
SOPUid is specified in the ( ) part,
%E = like %V but data is anonymized (new UID assigned),
%R = like %V but returned de-anonymized UID (reverse of %E),
%A = like %V but CRC32 of data is returned,
%QPxxxx,yyyy = item queried from patient db on patient ID (import converters only),
%QSxxxx,yyyy = queried from study db on patient ID and study UID (idem),
%QExxxx,yyyy = queried from series db on patient ID, study UID, and series UID (idem),
%QWxxxx,yyyy = dicom item queried from worklist db on accession number (idem),
%QXxxxx,yyyy = replace item from tab separated file aliasfileQX.txt (format: old\tnew\n)
%x, %y, %z = general purpose variables.
Each % variable can be appended with [first,last] to take a substring, i.e., %i[0,1] = first 2 characters of patientid, %i[,2] = last two characters of patient ID. A '^' may be appended to convert the result to uppercase, and a ~ to convert it to lowercase. For example, to change two VRs in each incoming image and reject any images acquired in 2002:
ImportConverter0 = set 0010,1001 to "my string and date: %d" ImportConverter1 = set 0010,1002 to "%V0010,0010^" ImportConverter2 = ifequal "%V0008,0020[0,3]", "2002"; destroy
The following list illustrates all importconverter ‘I’, exportconverter ‘E’, or both ‘IE’ commands available for scripting. The parser is not very flexible: stay close to the examples in terms of spacing and semicolons. The maximum allowed length of each command line (with % items expanded) is 512 characters.
IE | {command; command } | command block |
IE | write "my string" to file.txt | write file |
IE | append "date: %d" to file.txt | append to e.g. log file |
IE | nop | do nothing |
IE | nop any text %i | do nothing but log shows text |
IE | prefetch | delayed preread (cache) of patient from disk |
IE | preretrieve AE | delayed collect of entire patient from AE |
IE | call file | call file with ImportConverter strings, or script string |
I | file | call file with ImportConverter strings, or script string |
I | file.lua | call file with lua code |
I file.lua(command) call file with lua code, command --> command_line I file.lua(“command”) call file with lua code, command --> command_line E executable call executable
IE lua “chunk” execute lua chunk
IE lua:chunk execute lua chunk, must be last converter on line IE system command line make a call to windows or linux
IE return return from file, or same as stop
IE stop stop parsing this converter
IE silentstop stop parsing this converter, no message
I set xxxx,yyyy to "%V0010,0010" set VR (creates empty sequence if applicable) I set xxxx,yyyy to nil delete VR
I set PatientID to "%VPatientName"set/get VR by name
I set xxxx,yyyy format “%%d” to .. set VR formatted (%%s, %%d, %%x, %%f, %%g) I set xxxx,yyyy if "%V0010,0010" set VR if data
I set xxxx,yyyy.N/xxxx,yyyy to .. set VR in sequence (max one deep, may use names) I set xxxx,yyyy.*/xxxx,yyyy to .. Add VR to sequence (max one deep, may use names) I set x to "%QP0010,0010" set variable
I set y if "%x" set variable if data
I setifempty xxxx,yyyy to "hallo" set if VR empty (obsolete, not in sequences) I setifempty xxxx,yyyy if "%x" set if VR empty and %x not
I setifempty z to "hallo" set only if z empty
I setifempty z if "%i" set only if z empty and %i not
I delete xxxx,yyyy delete VR
I newuids replace all UIDS
I newuids except replace all UIDS except gggg,eeee|gggg,eeee or UID I newuids stage NAME replace all UIDS, but with separate keytable NAME I newuids stage #NAME replace UIDS, using a specific MD5 hash (one way) I olduids un-replace all UIDS
I olduids except un-replace all UIDS ex. gggg,eeee|gggg,eeee or UID I olduids stage NAME un-replace all UIDS, with separate keytable NAME I fixkodak change patient ID from kodak to NKI format
I unfixkodak change patient ID from NKI to kodak format
I crop x1,x2,y1,y2 crop image
I scrub +string/-string scrub vrs on type,private,grp,elemnt eg -SQ,7FE0 I tomono convert image to monochromo
IE save to filename.dcm save dicom image to file IE save bmp to filename.bmp save bitmap image to file IE save gif to filename.gif save gif image to file
IE save jpg to filename.jpg save jpeg image to file
IE save [bmp/gif/jpg] full syntax of above commands
MUSHU LOGINSTABLEUSD EXCHANGEOZONECHAIN LOGINCHECKR 交易所CRYPTO SCENARIOSETHEREUM EXPRESS LOGINSUPERWALK LOGINMMA APP
SHIBA TWITTERSHADOWCATS 交易所
CVXFPIS LOGINSHARE POWER TOKEN LOGINIS JD VANCE PRO CRYPTOCENTRA CRYPTO
IE save frame N to filename save dicom file of single frame of multiframe object IE mkdir directoryname make a directory (requires trailing / or \)
IE rm filename delete a file
I process with command received is processed by executable (max 256 chars) I destroy image not stored at all
I destroy2 same as destroy but not logged
I reject as destroy, but reports error to sending server
I storage MAG1 set preferred storage area
I compression CC[nn] recompress object with mode CC quality nn
I virtualserver N set preferred virtual servers (only for query/moves) I virtualservermask N set all preferred virtual servers (idem)
IE forward to AE see above
IE forward full syntax
HYGEIA LOGINATCG 交易所
[to AE|host:port] required, provide clauses in order
[org AE] calling optional, provide clauses in order
[dest AE] called optional, provide clauses in order
[channel N] optional (I only) to keep connection open: N={0..19 or * means distribute channels automatically}
[script cmd] script to process data; must be last IE forward patient to AE delayed forward of entire patient IE forward study to AE delayed forward of entire study IE forward series to AE delayed forward of entire series
IE forward [patient|study|series|image] forward command full syntax [compressed as xx] set compression
[date yyyymmdd-yyyymmdd] filter absolute series date range (any study) [now -ddd+ddd] filter series date range from now (any study)
[age -ddd+ddd] filter series date range from passed series (any study)
[modality mm] filter modality (any study)
[imagetype xxxx] filter image type (this study)
[seriesdesc xxxx] filter series description (this study)
[study xxx] filter studyUID (any series)
[series xxx] filter seriesUID (any study)
[sop xxxx] filter sop (any study)
[split N/M] send subset N(0..M-1) of 1/M images (1.5.0)
[after NN] collect delay in seconds from last image
XRN EXCHANGESCHAMPIGNONS OF ARBORETHIA APP
[script “....”] must be last: script to run on sent objects (“ optional) maximum script length is 400 characters
IE get [patient|study|series|image] get command full syntax [date yyyymmdd-yyyymmdd] (see above)
[now -ddd+ddd] [age -ddd+ddd] [modality mm] [imagetype xxxx] [seriesdesc xxxx] [study xxx] [series xxx] [sop xxxx] [after NN]
REDPANDA EARTH EXCHANGESNRC LOGINETHERBACK EXCHANGESSPHYNXFI EXCHANGES
IE delete [patient|study|series|image] delete command full syntax [date yyyymmdd-yyyymmdd] (see above)
[now -ddd+ddd] [age -ddd+ddd] [modality mm] [imagetype xxxx] [seriesdesc xxxx] [study xxxx] [series xxxx] [sop xxxx] [after NN]
IE submit [patient|study|series|image] secure ftp submit command full syntax [target xxxx] username@machine:folder [password xxxx]
[after NN]
[study xxx] filter studyUID (default current)
[series xxx] filter seriesUID (default current)
[sop xxxx] filter sop (default current)
[script “....”] must be last: script to anonymize (max 400 chars) Note: calls submit.cq for every image if it exists.
IE submit2 [patient|study|series|image] submit using any tool command full syntax [target xxxx] substituted in command line below
[command xxxx] command line string with %s=filename %s=target [command [xxxx]] embed in [] to allow spaces in command line [after NN]
[study xxx] filter studyUID (default current)
[series xxx] filter seriesUID (default current)
[sop xxxx] filter sop (default current)
[script “....”] must be last: script to anonymize (max 400 chars) Note: calls submit.cq for every image if it exists.
IE merge study series in a study are merged [modality mm] (any study)
[seriesdesc xxxx] (this study) [after NN]
[script “....”] importconverter run on merged objects (max 400) IE process [patient|study|series|image]
[after NN]
[by command/file.lua(arguments)] the executable command or lua file (passed
command_line as variable) is executed when reception of the patient etc is complete (command is max 360 characters).
IE testmode .1 controls appending of internal DICOM filename
IE copy file to file see above
IE defer defer ExportConverter until another time IE ifnotempty "%i"; command if filled then command
IE ifempty "%V0010,0010"; nop if "" then .. IE ifequal "string", "string2"; nop test equal IE ifnotequal "string", "string2"; nop test not equal
IE ifmatch "string", "substring"; nop test match (allows substring = x*, *x, and *x*) IE ifnotmatch "string", "substring" test not match (allows substring = x*, *x, and *x*) IE ifnumequal "string", "string2" test numeric
IE ifnumnotequal "string", "string2" test numeric IE ifnumgreater "string", "string2" test numeric IE ifnumless "string", "string2" test numeric
IE ifnotempty "%i"; {nop; nop; } {} block (note ‘;’ use!)
IE ifequal "%V0008,0020[0,3]", "2002"; substring to test year IE between "9", "17"; defer; test time in hours
Script strings are entered in dicom.ini as follows: [scripts]
Test = nop test
Here is a real-life example of a useful exportconverter script in SERVER1:
exportconverters = 1
exportconverter0 = ifequal "%u","SERVER2"; stop; between "9", "17"; defer; forward to SERVER2
This script uses the commands 'ifequal "%u","SERVER2"; stop;' to ignore all data with calling AE of 'SERVER2'. This will avoid any data from SERVER2 to be sent back to SERVER2 causing a potential loop. The commands "between 9 and 17; defer" cause the converter to wait until after 17:00 before subsequent commands are processed using the retrying mechanism. The last command forwards the data to SERVER2. Having a similar line in SERVER2 forwarding
to SERVER1 will cause both servers to synchronize after 17:00 without a loop.
Here is a much more elaborate sample that when it receives an RTPLAN for machine A2 will forward the associated RTSTRUCT and CT to machine A2:
exportconverters = 1
exportconverter0 = ifnotequal "%m", "RTPLAN"; stop;
ifnotequal "%V*300a,00b2[0,1]", "A2"; stop; forward to XVI_A2;
get study modality CT from NKIPACS;
get study modality RTSTRUCT sop %V/300c,0060.0/0008,1155 from NKIPACS;
forward study series %V(/300c,0060/0008,1155)/3006,0010/3006,0012/3006,0014/0020,000e to XVI_A2;
forward study modality RTSTRUCT sop %V/300c,0060.0/0008,1155 to XVI_A2
WRAPPED STAR LOGINI BOUGHT $1 WORTH OF BITCOIN ON CASH APPSTRAINER COINBASEIMPERIUM EMPIRES
BEST CRYPTO CASINO 2023UNILOCK NETWORK 交易所
DEFIVILLE EXCHANGESPARSIQ LOGIN
TWS APPYEARN FINANCE BIT2
AL MUSK LOGINHOOKED PROTOCOL 交易所
AIDOSMARKET EXCHANGEPAC PROTOCOL 交易所
SREUR EXCHANGESZOOPIA EXCHANGE
SERV EXCHANGESXRUNE EXCHANGE
Conquest addition.
FUTURE 交易所BABY CAT INU APP
PDF LOGINSCAT EXCHANGES
DIBIGLOBAL LOGINCONNECT EXCHANGES
APRONNETWORK EXCHANGEFKX LOGINKINBA APPWANA EXCHANGE
CLUBE ATLÉTICO MINEIRO FAN TOKEN APPGSKY 交易所JAMES ALTUCHER CRYPTO PREDICTIONCRYPTO WALLET APP DEVELOPMENTKEKE EXCHANGEPIZZAUSDE EXCHANGES
HOW TO BYPASS COINBASE ID VERIFICATIONSHIBA ELON 交易所
KNIGHT LOGIN
--[[
#Conquest DICOM server scripting overview
Brief demo of _all_ scripting options in **Conquest Dicom Server**. If you run this script from **ZeroBrane Studio**, you can put breakpoints on any line, single step, inspect data with the mouse, and display arbitrary data and enter arbitrary commands to the DICOM Server in the `Local console` window.
In version 1.4.19 for Windows the following modules are embedded in the executable:
-- `require('socket')`
-- `require('pack')`
-- and string:reshape(string, i, j) is embedded
]]
-- for demo fill the global variable Data which normally contains
-- the incoming DICOM object. You can read from disk or from the
-- server using a PatientID:SOPInstanceUID format. For reading from
-- virtual server use 'StudyUID\SeriesUID\SOPInstanceUID format.
-- readdicom('c:\\t.dcm') -- from disk readdicom('0009703828:1.3.46.670589.5.2.10.2156913941.892665340.475317')
-- Note that in this demo Data is undefined until after this call
-- I.e., Data:Read() is not allowed as first call
-- association info available from lua:
print(Association.Calling, Association.Called, Association.Thread, Association.ConnectedIP)
-- all counters available from lua: print('------ All counters --------') print(Global.StartTime) print(Global.TotalTime) print(Global.LoadTime) print(Global.ProcessTime)
print(Global.SaveTime) print(Global.ImagesSent) print(Global.ImagesReceived) print(Global.ImagesSaved) print(Global.ImagesSaved) print(Global.ImagesForwarded) print(Global.ImagesExported) print(Global.ImagesCopied) print(Global.IncomingAssociations) print(Global.EchoRequest) print(Global.C_Find_PatientRoot) print(Global.C_Move_PatientRootNKI) print(Global.C_Move_PatientRoot) print(Global.C_Find_StudyRoot) print(Global.C_Move_StudyRootNKI) print(Global.C_Move_StudyRoot) print(Global.C_Find_PatientStudyOnly) print(Global.C_Find_ModalityWorkList) print(Global.C_Move_PatientStudyOnlyNKI) print(Global.C_Move_PatientStudyOnly) print(Global.UnknownRequest) print(Global.CreateBasicFilmSession) print(Global.DeleteBasicFilmSession) print(Global.ActionBasicFilmSession) print(Global.SetBasicFilmSession) print(Global.CreateBasicFilmBox) print(Global.ActionBasicFilmBox) print(Global.SetBasicFilmBox) print(Global.DeleteBasicFilmBox) print(Global.SetBasicGrayScaleImageBox) print(Global.SetBasicColorImageBox) print(Global.GuiRequest) print(Global.ImagesToGifFromGui) print(Global.ImagesToDicomFromGui) print(Global.ExtractFromGui) print(Global.QueryFromGui) print(Global.DeleteImageFromGui) print(Global.DeletePatientFromGui) print(Global.DeleteStudyFromGui) print(Global.DeleteStudiesFromGui) print(Global.DeleteSeriesFromGui) print(Global.MovePatientFromGui) print(Global.MoveStudyFromGui) print(Global.MoveStudiesFromGui) print(Global.MoveSeriesFromGui) print(Global.AddedFileFromGui) print(Global.DumpHeaderFromGui) print(Global.ForwardFromGui) print(Global.GrabFromGui) print(Global.DatabaseOpen) print(Global.DatabaseClose) print(Global.DatabaseQuery) print(Global.DatabaseAddRecord) print(Global.DatabaseDeleteRecord) print(Global.DatabaseNextRecord) print(Global.DatabaseCreateTable) print(Global.DatabaseUpdateRecords) print(Global.QueryTime)
print(Global.SkippedCachedUpdates) print(Global.UpdateDatabase) print(Global.AddedDatabase) print(Global.NKIPrivateCompress) print(Global.NKIPrivateDecompress) print(Global.DownSizeImage) print(Global.DecompressJpeg) print(Global.CompressJpeg) print(Global.DecompressJpeg2000) print(Global.CompressJpeg2000) print(Global.DecompressedRLE) print(Global.DePlaned) print(Global.DePaletted) print(Global.RecompressTime) print(Global.gpps) print(Global.gppstime)
-- all configuration items are available from lua (read/write) print(Global.NoDicomCheck)
print(Global.DebugLevel) print(Global.ThreadCount) print(Global.OpenThreadCount) print(Global.EnableReadAheadThread) print(Global.WorkListMode) print(Global.StorageFailedErrorCode) print(Global.FailHoldOff) print(Global.RetryDelay) print(Global.QueueSize) print(Global.ForwardCollectDelay) print(Global.CacheVirtualData) print(Global.gJpegQuality) print(Global.gUseOpenJpeg) print(Global.FixKodak) print(Global.NumIndexing) print(Global.DoubleBackSlashToDB) print(Global.UseEscapeStringConstants) print(Global.EnableComputedFields) print(Global.FileCompressMode) print(Global.RootConfig) print(Global.BaseDir) print(Global.DGATEVERSION) print(Global.ConfigFile) print(Global.DicomDict) print(Global.AutoRoutePipe) print(Global.AutoRouteExec) print(Global.DroppedFileCompression) print(Global.IncomingCompression) print(Global.TransmitCompression) print(Global.DefaultNKITransmitCompression) print(Global.ArchiveCompression) print(Global.TestMode) print(Global.StatusString)
print('------ Set a config item --------')
Global.DebugLevel = 4
print('----- Read any dicom.ini item -----') section = 'sscscp'; item = 'TCPPort'; default='';
print(gpps(section, item, default))
-- all command items are available from lua (read only)
print('------ testing debug log (typically not shown) --------', 1234) debuglog('Command priority', Command.Priority)
-- set Data storage
print('------ test setting storage --------') Data.Storage = 'MAG2'
print('You can only read/write storage in an import converter', Data.Storage);
-- read/write data, create sequences, and write into sequences (if [] not passed, [0] is assumed) print('------ test read/write Data --------')
Data.PatientName = Data.PatientID Data.ReferencedStudySequence = {}
print('Number of elements in sequence', #Data.ReferencedStudySequence); print('This is a sequence: ', Data.ReferencedStudySequence); Data.ReferencedStudySequence[0].StudyInstanceUID = Data.StudyInstanceUID Data.ReferencedStudySequence.StudyInstanceUID = Data.StudyInstanceUID Data.ReferencedStudySequence[1].StudyInstanceUID = Data.StudyInstanceUID print(#Data.ReferencedStudySequence);
print('This is a sequence item: ', Data.ReferencedStudySequence[0].StudyInstanceUID);
-- list items in Object, returns names,types,groups,elements print(Data.ReferencedStudySequence[1]:ListItems())
-- Delete sequence item:
-- Object:DeleteFromSequence(name, item)
-- Array:Delete(item) deletefromsequence(Data.ReferencedStudySequence, 0); Data.ReferencedStudySequence[1]:Delete()
-- delete items
print('------ test delete item --------')
Data.ReferencedStudySequence = nil
print('This was a sequence: ', Data.ReferencedStudySequence);
-- inspect dictionary (results in 16, 32 vs PatientID) print(dictionary('PatientID'))
print(dictionary(16, 32))
-- inspect sql definition (database, row) results in 16, 32 PatientID 64 SQL_STR DT_STR) print(get_sqldef(0, 0))
-- send a script to conquest
print('------ test conquest script call --------')
script('nop this is an ImportConverter script') -- only works when Data defined Data:Script('nop this is an ImportConverter script running on a specified DICOM object')
-- send a servercommand to conquest and read its result print('------ test conquest command call --------') print(servercommand('display_status:'))
servercommand('display_status:', 'cgibinary') -- for web interface; also allows value 'cgihtml' or <filename to upload and >filename to download or 'binary' without processing
-- run executable in the background (avoids window opened by os.execute) system('dgate.exe -?')
-- get an item from ACRNEMA.MAP
print('------ test reading ACRNEMA.MAP --------') print(get_amap(0))
-- results in 'CONQUESTSRV1 127.0.0.1 5678 un'
-- remap UIDs (in 1.4.17) print(changeuid('12jgkfjgfkgjax', 'aapnootmies')) print(changeuidback('aapnootmies')) print(changeuid('1.1'))
print(changeuidback('1.2.826.0.1.3680043.2.135.734877.42238624.7.1359125302.31.0'))
print(genuid())
-- results in:
-- [CONQUESTSRV1] aapnootmies
-- [CONQUESTSRV1] 12jgkfjgfkgjax
-- [CONQUESTSRV1] 1.2.826.0.1.3680043.2.135.734877.42238624.7.1359125302.31.0
-- [CONQUESTSRV1] 1.1
-- [CONQUESTSRV1] .... a new uid here ....
-- To remap all uids use script('newuids') and reverse (1.4.17) with script('olduids')
-- staged remap UIDs (in 1.4.19) print(changeuid('12jgkfjgfkgjax', 'aapnootmies', 'stage1')) print(changeuidback('aapnootmies', 'stage1')) print(changeuid('1.1', 'stage1'))
print(changeuidback('1.2.826.0.1.3680043.2.135.734877.42238624.7.1359125302.31.0', 'stage1'))
print(genuid())
– MD5 has based uid remapping (forward only): print(changeuid('1.1', '#stage1'))
-- query the local database (also possible from CGI interface, if the database is setup in the CGI dicom.ini) print('------ test quering a database --------')
print(dbquery('DICOMPatients', 'PatientNam', 'PatientID LIKE \'2%\'')[1][1])
-- set and get pixels in the current image or any loaded image print('------ test reading and writing pixels --------')
x=0; y=0; frame=0; print(getpixel(x, y, frame));
setpixel(x, y, frame, getpixel(x, y, frame)*2+10); print(getpixel(x, y, frame)); print(Data:GetPixel(x, y, frame))
-- set and get rows and colums in the image
print('------ test reading and writing rows and columns --------') a = getrow(Data.Rows / 2)
a = Data:GetRow(Data.Rows / 2) print(a[1], a[2], a[3], a[4], a[128]);
setcolumn(Data.Columns / 2, frame, a) Data:SetColumn(Data.Columns / 3, frame, a)
-- get / set image as binary string, also allow efficient binary image conversion (1.4.17) a = getimage(frame); a = Data:GetImage(frame)
setimage(frame, a) Data:SetImage(frame, a)
-- get / set image as binary string with floats (1.4.17a)
-- read a from a floating point image, i.e., it has 4 bytes per pixel, scale passed
-- setimage(frame, a, 1000); Data:SetImage(frame, a, 1000)
-- copy a dicom object
local c = Data:Copy() c:Write('c:\\copy.dcm') c:free()
-- compress a dicom object local c = Data:Compress('jk') c:Write('c:\\compressed_jk.dcm') c:free()
-- create/read/write/destroy a dicom object
print('------ test create/read/write dicom object --------') a = newdicomobject()
a = DicomObject:new() -- preferred notation in 1.4.17 a.PatientID = 'test'
writedicom(a, 'c:\\file1.dcm') b = newdicomobject() readdicom(b, 'c:\\file1.dcm') writeheader(b, 'c:\\file1.txt') a:Write('c:\\file2.dcm') a:Read('c:\\file2.dcm')
-- document read here a:Dump('c:\\file2.txt') a:GetPixel(x, y, z) a:SetPixel(x, y, z, value) a:GetRow(x, y, z) a:SetRow(x, y, z, table) a:GetColumn(x, y, z) a:SetColumn(x, y, z, table)
deletedicomobject(a) -- not required: will be freed automatically
-- a:free() -- also allowed in 1.4.17:
-- add json object to dicom object, where the json_string can have elements by name or number. local c = Data:Copy(json_string)
-- convert json object to dicom object, where the json_string can have elements by name or number. local c = DicomObject:new(json_string)
-- query a dicomserver (returns a dicomobjectarray) print('------ test query a dicom server --------')
b=newdicomobject(); b.PatientName = '*'; a=dicomquery('CONQUESTSRV1', 'PATIENT', b); print ('First query result has this patientname:', a[0].PatientName);
-- deletedicomobject(a) -- not required: will be freed automatically
-- delete data from local dicomserver (use with care) print('------ delete from dicom server --------')
b=newdicomobject(); b.PatientID = 'hopedoesntexist'; dicomdelete(b); local threadno_forprogressinfo=123
b=newdicomobject(); b.PatientID = 'hopedoesntexist'; newdicomdelete(b, threadno_forprogressinfo);
-- modify data from local dicomserver (use with care) local threadno_forprogressinfo=123
local copy=1
b=newdicomobject(); b.PatientID = 'hopedoesntexist'; newdicommodify(b, 'nop', threadno_forprogressinfo, copy);
-- create a dicomobjectarray
print('------ test creating dicom array --------') a=newdicomarray(); a[0].PatientID='a'; a[1].PatientID='b';
-- in 1.4.17 also: a = DicomObject:newarray()
-- read the filename of a dropped file if any print('------ test Filename variable --------') print('Is there a file dropped?', Filename)
-- access to long and sequence VRs
print('------ test reading / writing long VRs --------') y = Data:GetVR(0x7fe0, 0x10);
print('Length of y', #y); y = getvr(0x7fe0, 0x10); setvr(0x7fe0, 0x10, y);
Data:SetVR(0x7fe0, 0x10, y);
-- where y is either a table starting at 1, or a dicomobjectarray for a sequence
-- y can also be a json string; can be used to fill sequence
-- In 1.4.17 these command can also return a more efficient binary string: y = Data:GetVR(0x7fe0, 0x10, true);
Data:SetVR(0x7fe0, 0x10, y);
-- memory allocation debugging
print('------ memory alloc check NOTE: CALL IS NOT THREAD SAFE --------') print(heapinfo());
-- move
print('------ testing a C-MOVE --------') AE = 'CONQUESTSRV1';
b=newdicomobject(); b.PatientName = 'HEAD EXP2'; b.QueryRetrieveLevel = 'STUDY'; dicommove('CONQUESTSRV1', AE, b);
b=newdicomobject(); b.PatientName = 'HEAD EXP2'; b.QueryRetrieveLevel = 'STUDY'; dicommove('CONQUESTSRV1', AE, b, 0, 'print(Global.StatusString)'); b=newdicomobject(); b.PatientName = 'HEAD EXP2'; b.QueryRetrieveLevel = 'PATIENT'; dicommove('CONQUESTSRV1', AE, b, 1);
--get
b=newdicomobject(); b.PatientName = 'HEAD EXP2'; b.QueryRetrieveLevel = 'STUDY'; array=dicomget('CONQUESTSRV1', 'IMAGE', b);
--read array of objects
b=newdicomobject(); b.PatientName = 'HEAD EXP2'; array=dicomread(b);
-- advanced move; 1.5.0 up
print('------ testing advanced C-MOVE; example setting max sent slices --------') b=newdicomobject();
c=newdicomobject(); c.ConquestMaxSlices=1
b.PatientName = 'HEAD EXP2'; b.QueryRetrieveLevel = 'PATIENT'; dicommove('CONQUESTSRV1', AE, b, 1, c);
– convert DICOM object to string print(c:Serialize()) -- lua format print(c:Serialize(true)) – json format
print(c:Serialize(true, true)) – json format with pixel data print(c:Serialize(true, true, true)) – dicomweb json format with pixel data
-- sql
print('------ testing an SQL statement --------')
sql("CREATE TABLE UserTable (CounterID varchar(99), Val integer)");
sql("INSERT INTO UserTable (CounterID,Val) VALUES ('CT',1) ON DUPLICATE KEY UPDATE Val=Val+1");
-- sleep, delay in ms sleep(1000)
-- mkdir, make folder recursively mkdir('c:\\temp\\test')
-- enter object into server: x = DicomObject:new()
x:Read('c:\\t.dcm'); x:AddImage() -- or addimage(x)
-- special script command 'retry'
print('------ testing retry script command --------')
script('retry') -- when used in RejectedImageWorkListConverter0 and RejectedImageConverter0; will re-attempt to store the object after the script is done; in 1.4.19 also retry()
-- special script command 'defer'
print('------ testing defer script command --------')
script('defer') -- when used in an ExportConverter, the convert will re-attempt to process or forward the object later
POLY SWARM CRYPTO
This file is placed in the same directory as the executable (e.g., c:\dicomserver). It specifies the configuration of the SQL database used to store IOD module attributes for Query/Retrieve operations. The Conquest DICOM server generates (and overwrites) it automatically upon first installation (i.e., when dicom.ini does not exist). Editing this file is not necessary, in 1.5.0 the name of the previous fields ‘Rows’ and ‘Colums’ in the image database we changed to ‘QRows’ and ‘QColumns". It is possible to check the syntax of this file for errors using the "List Database Layout" button on the "Maintenance" page of the Conquest DICOM server. Note that the database definition defines a copy of the PatientID in both the series and the image table. This is done to allow improved query speed.
From version 1.4.0 on, the contents of this file depend on the selected database driver upon installation (when dicom.ini does not exist), where the built in dBaseIII driver uses a non- normalized version of the database (not listed here), and the others use the file as listed here.
Implementing changed versions of this file requires a full regeneration of the database. Without full regeneration, the server will not function correctly! Removing fields from this database may affect the DICOM server user interface operation.
When adding database fields leave the first and last field in place; they are the primary key and the link to the higher database. The server fails if they are removed or moved. To force case insensitive queries (1.4.19c up), replace 'DT_STR' with 'DT_ISTR' on relevant columns. This can be done without regenerating the database.
The worklist database has an extra column with HL7 tags used for translating HL7 data to a dicom worklist. These tags can be changed at any time without regenerating the database, restarting the server suffices to use the new tags.
Push "Clear worklist database" on the installation page of the GUI to create a fresh worklist database.
/*
# DICOM Database layout
# Example version for all SQL servers (mostly normalized)
#
# (File DICOM.SQL)
# ** DO NOT EDIT THIS FILE UNLESS YOU KNOW WHAT YOU ARE DOING **
#
# Version with modality moved to the series level and EchoNumber in image table
# Revision 3: Patient birthday and sex, bolus agent, correct field lengths
# Revision 4: Studymodality, Station and Department in study
# Manufacturer, Model, BodyPart and Protocol in series
# Acqdate/time, coil, acqnumber, slicelocation and pixel info in images
# Notes for revision 4:
# DepartmentName in study (should officially be in series, but eFilm expects it in study)
# StationName is in study (should officially be in series, but more useful in study)
# Revision 5: Added patientID in series and images for more efficient querying
# Revision 6: Added frame of reference UID in series table
# Revision 7: Added ImageType in image table, StudyModality to 64 chars, AcqDate to SQL_C_DATE
# Revision 8: Denormalized study table (add patient ID, name, birthdate) to show consistency problems
# Revision 10: Fixed width of ReceivingCoil: to 16 chars
# Revision 13: Added ImageID to image database
# Revision 14: Added WorkList database with HL7 tags
# Revision 16: Moved Stationname and InstitutionalDepartmentName to series table
# Revision 17: EchoNumber, ReqProcDescription to 64 characters; StudyModality, EchoNumber, ImageType to DT_MSTR; use
# Institution instead of InstitutionalDepartmentName');
# Revision 18: DT_STR can now be replaced by DT_ISTR to force case insensitive searches
# Revision 19: Use QRows and QColumns as field names
#
# 5 databases need to be defined:
#
# *Patient*
# *Study*
# *Series*
# *Image*
# *WorkList*
#
#
# The last defined element of Study is a link back to Patient
# The last defined element of Series is a link back to Study
# The last defined element of Image is a link back to Series
#
#
# Format:
# { Group, Element, Column Name, Column Length, SQL-Type, DICOM-Type }
*/
*Patient*
{
}
*Study*
{
}
*Series*
{
{ 0x0010, 0x0020, "PatientID", 64, SQL_C_CHAR, DT_STR },
{ 0x0010, 0x0010, "PatientName", 64, SQL_C_CHAR, DT_STR },
{ 0x0010, 0x0030, "PatientBirthDate", 8, SQL_C_DATE, DT_DATE },
{ 0x0010, 0x0040, "PatientSex", 16, SQL_C_CHAR, DT_STR }
{ 0x0020, 0x000d, "StudyInstanceUID", 64, SQL_C_CHAR, DT_UI },
{ 0x0008, 0x0020, "StudyDate", 8, SQL_C_DATE, DT_DATE },
{ 0x0008, 0x0030, "StudyTime", 16, SQL_C_CHAR, DT_TIME },
{ 0x0020, 0x0010, "StudyID", 16, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x1030, "StudyDescription", 64, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0050, "AccessionNumber", 16, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0090, "ReferPhysician", 64, SQL_C_CHAR, DT_STR },
{ 0x0010, 0x1010, "PatientsAge", 16, SQL_C_CHAR, DT_STR },
{ 0x0010, 0x1030, "PatientsWeight", 16, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0061, "StudyModality", 64, SQL_C_CHAR, DT_MSTR },
{ 0x0010, 0x0010, "PatientName", 64, SQL_C_CHAR, DT_STR },
{ 0x0010, 0x0030, "PatientBirthDate", 8, SQL_C_DATE, DT_DATE },
{ 0x0010, 0x0040, "PatientSex", 16, SQL_C_CHAR, DT_STR }
{ 0x0010, 0x0020, "PatientID", 64, SQL_C_CHAR, DT_STR }
{ 0x0020, 0x000e, "SeriesInstanceUID", 64, SQL_C_CHAR, DT_UI },
}
*Image*
{
}
{ 0x0020, 0x0011, "SeriesNumber", 12, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0021, "SeriesDate", 8, SQL_C_DATE, DT_DATE },
{ 0x0008, 0x0031, "SeriesTime", 16, SQL_C_CHAR, DT_TIME },
{ 0x0008, 0x103e, "SeriesDescription", 64, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0060, "Modality", 16, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x1010, "StationName", 16, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0080, "Institution", 64, SQL_C_CHAR, DT_STR },
{ 0x0018, 0x5100, "PatientPosition", 16, SQL_C_CHAR, DT_STR },
{ 0x0018, 0x0010, "ContrastBolusAgent", 64, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0070, "Manufacturer", 64, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x1090, "ModelName", 64, SQL_C_CHAR, DT_STR },
{ 0x0018, 0x0015, "BodyPartExamined", 64, SQL_C_CHAR, DT_STR },
{ 0x0018, 0x1030, "ProtocolName", 64, SQL_C_CHAR, DT_STR },
{ 0x0020, 0x0052, "FrameOfReferenceUID", 64, SQL_C_CHAR, DT_UI },
{ 0x0010, 0x0020, "SeriesPat", 64, SQL_C_CHAR, DT_STR },
{ 0x0020, 0x000d, "StudyInstanceUID", 64, SQL_C_CHAR, DT_UI }
{ 0x0008, 0x0018, "SOPInstanceUID", 64, SQL_C_CHAR, DT_UI },
{ 0x0008, 0x0016, "SOPClassUID", 64, SQL_C_CHAR, DT_UI },
{ 0x0020, 0x0013, "ImageNumber", 12, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0023, "ImageDate", 8, SQL_C_DATE, DT_DATE },
{ 0x0008, 0x0033, "ImageTime", 16, SQL_C_CHAR, DT_TIME },
{ 0x0018, 0x0086, "EchoNumber", 64, SQL_C_CHAR, DT_MSTR },
{ 0x0028, 0x0008, "NumberOfFrames", 12, SQL_C_CHAR, DT_STR },
{ 0x0008, 0x0022, "AcqDate", 8, SQL_C_DATE, DT_DATE },
{ 0x0008, 0x0032, "AcqTime", 16, SQL_C_CHAR, DT_TIME },
{ 0x0018, 0x1250, "ReceivingCoil", 16, SQL_C_CHAR, DT_STR },
{ 0x0020, 0x0012, "AcqNumber", 12, SQL_C_CHAR, DT_STR },
{ 0x0020, 0x1041, "SliceLocation", 16, SQL_C_CHAR, DT_STR },
{ 0x0028, 0x0002, "SamplesPerPixel", 5, SQL_C_CHAR, DT_UINT16 },
{ 0x0028, 0x0004, "PhotoMetricInterpretation", 16, SQL_C_CHAR, DT_STR },
{ 0x0028, 0x0010, "QRows", 5, SQL_C_CHAR, DT_UINT16 },
{ 0x0028, 0x0011, "QColumns", 5, SQL_C_CHAR, DT_UINT16 },
{ 0x0028, 0x0101, "BitsStored", 5, SQL_C_CHAR, DT_UINT16 },
{ 0x0008, 0x0008, "ImageType", 128, SQL_C_CHAR, DT_MSTR },
{ 0x0054, 0x0400, "ImageID", 16, SQL_C_CHAR, DT_STR },
{ 0x0010, 0x0020, "ImagePat", 64, SQL_C_CHAR, DT_STR },
{ 0x0020, 0x000e, "SeriesInstanceUID", 64, SQL_C_CHAR, DT_UI }
*WorkList*
{
{ 0x0008, 0x0050, "AccessionNumber", 16, SQL_C_CHAR, DT_STR, "OBR.3" },
{ 0x0010, 0x0020, "PatientID", 64, SQL_C_CHAR, DT_STR, "PID.4" },
{ 0x0010, 0x0010, "PatientName", 64, SQL_C_CHAR, DT_STR, "PID.5" },
{ 0x0010, 0x0030, "PatientBirthDate", 8, SQL_C_DATE, DT_DATE, "PID.7" },
{ 0x0010, 0x0040, "PatientSex", 16, SQL_C_CHAR, DT_STR, "PID.8" },
{ 0x0010, 0x2000, "MedicalAlerts", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0010, 0x2110, "ContrastAllergies", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0020, 0x000d, "StudyInstanceUID", 64, SQL_C_CHAR, DT_UI, "---" },
{ 0x0032, 0x1032, "ReqPhysician", 64, SQL_C_CHAR, DT_STR, "OBR.16" },
{ 0x0032, 0x1060, "ReqProcDescription", 16, SQL_C_CHAR, DT_STR, "OBR.4.1" },
{ 0x0040, 0x0100, "--------", 0, SQL_C_CHAR, DT_STARTSEQUENCE, "---" },
{ 0x0008, 0x0060, "Modality", 16, SQL_C_CHAR, DT_STR, "OBR.21" },
{ 0x0032, 0x1070, "ReqContrastAgent", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0001, "ScheduledAE", 16, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0002, "StartDate", 8, SQL_C_DATE, DT_DATE, "OBR.7.DATE" },
{ 0x0040, 0x0003, "StartTime", 16, SQL_C_CHAR, DT_TIME, "OBR.7.TIME" },
{ 0x0040, 0x0006, "PerfPhysician", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0007, "SchedPSDescription", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0009, "SchedPSID", 16, SQL_C_CHAR, DT_STR, "OBR.4" },
{ 0x0040, 0x0010, "SchedStationName", 16, SQL_C_CHAR, DT_STR, "OBR.24" },
{ 0x0040, 0x0011, "SchedPSLocation", 16, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0012, "PreMedication", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0400, "SchedPSComments", 64, SQL_C_CHAR, DT_STR, "---" },
{ 0x0040, 0x0100, "---------", 0, SQL_C_CHAR, DT_ENDSEQUENCE, "---" },
{ 0x0040, 0x1001, "ReqProcID", 16, SQL_C_CHAR, DT_STR, "OBR.4.0" },
{ 0x0040, 0x1003, "ReqProcPriority", 64, SQL_C_CHAR, DT_STR, "OBR.27 }
}
WORM EXCHANGES
This file is placed in the same directory as the executable (e.g., c:\dicomserver). It specifies the configuration of the ACR-NEMA to IP address and port map, used for Query/Retrieve operations. Most DICOM servers and applications will NOT communicate with the Conquest DICOM server unless they have been correctly added to this list and this server has been made known to them. This file also specifies the type of compression that will be proposed for outgoing connections. The accepted values are the same as for DroppedFileCompression in dicom.ini, with the exception that transmission of dicom objects in "as" and "nj" modes is not correctly implemented and should only be used with NKI clients or the Conquest DICOM server.
Upon installation, an empty version of this file is created automatically (the installation program will NOT overwrite this file if it exists). Edit the contents of this file through the "Known DICOM providers" page of the Conquest DICOM server. Do not change the file header.
It is possible to check the syntax of this file for errors using the "List DICOM providers"
button on the "Maintenance" page of the Conquest DICOM server.
It is possible to test communication with other DICOM servers (that support the Query/Move functionality, i.e., image servers) through the "Query / Move" page of the Conquest DICOM server.
Conquest addition: this file supports a simple wild-card mechanism. The AE, host name and IP port may all end on a *. The * part of the AE is copied into the host name and/or IP port without change. In the following example any application with an AE of "V" followed by its IP number or host name will be allowed to communicate through port 1234.
The wildcard option is highly useful to let a group of, e.g., viewer applications or servers communicate without having to configure each of them individually in the server. Add normal entries above wildcard entries as they may conflict in name.
Note that lua scripts may read this file and interpreted addition columns. File association.lua shows how to add a column to enable multithreaded sends, and to enable removal of patient names.
/* **********************************************************
* *
DICOM AE (Application entity) -> IP address / Port map *
(This is file ACRNEMA.MAP) *
* *
All DICOM systems that want to retrieve images from the *
Conquest DICOM server must be listed here with correct *
AE name, (IP adress or hostname) and port number. The *
first entry is the Conquest server itself. The last ones *
with * show wildcard mechanism. Add new entries above. *
* *
The syntax for each entry is : *
AE <IP adress|Host name> port number compression *
* *
For compression see manual. Values are un=uncompressed; *
ul=littleendianexplicit,ub=bigendianexplicit,ue=both *
j2=lossless jpeg;j3..j6=lossy jpeg;n1..n4=nki private *
js =lossless jpegLS; j7=lossy jpegLS *
jk =lossless jpeg2000;jl=lossy jpeg2000 *
J3NN..j6NN, JLNN or J7NN overrides quality factor to NN% *
* *
********************************************************** */
CONQUESTSRV1 | 127.0.0.1 | 5678 | un |
V* | * | 1234 | un |
W* | * | 666 | un |
S* | * | 5678 | un |
Note that acrnema.map does not check for double entries yet, only the first entry is used, therefore do not put anything below entries with a *. Lua scripts (see association.lua) can add additional columns after the four predefined ones, e.g. to comfigute multithreaded moves.
FREE LOGIN
This file is placed in the same directory as the executable (e.g., c:\dicomserver). It specifies the configuration of the SSC-SCP engine. This file can also be used to selectively reject other SOP classes, as well as provide security for incoming AE’s. The Conquest DICOM server generates it automatically upon installation. A copy of this file is present in the TEMP directory. This latter copy is automatically removed when closing the server. From version 1.4.0, GEMRStorage and GECTStorage are disabled (using ‘#’), thereby forcing GE scanners to transmit standard DICOM images that other viewers can handle. From version 1.4.2 up, JPEG transfer syntaxes are enabled for incoming connections if JPEG support is configured as ON. To filter incoming requests from unknown AE addresses start adding RemoteAE lines, not forgetting to add the server AE itself as well.
#
# DICOM Application / sop / transfer UID list.
#
# This list is used by the CheckedPDU_Service ( "filename" ) service
# class. All incoming associations will be verified against this
# file.
#
# Revision 2: disabled GEMRStorage and GECTStorage
# Revision 3: extended with new sops and with JPEG transfer syntaxes
# Revision 4: added Modality Worklist query
# Revision 5: Erdenay added UIDS for CTImageStorageEnhanced, RawDataStorage,
# SpatialRegistrationStorage, SpatialFiducialsStorage, DeformableSpatialRegistrationStorage,
# SegmentationStorage, SurfaceSegmentationStorage, RTIonPlanStorage, RTIonBeamsTreatmentRecordStorage
# Revision 6: X-RayRadiationDoseSR, BreastTomosynthesisImageStorage and others
# Revision 7: JpegLS and BigEndianExplicit enabled
# Revision 8: Added C-GET
#
#None none RemoteAE
#None none LocalAE
#DICOM 1.2.840.10008.3.1.1.1 application
Verification 1.2.840.10008.1.1 sop
RETIRED_StoredPrintStorage | 1.2.840.10008.5.1.1.27 | sop | |
RETIRED_HardcopyGrayscaleImageStorage | 1.2.840.10008.5.1.1.29 | sop |
RETIRED_HardcopyColorImageStorage | 1.2.840.10008.5.1.1.30 | sop |
ComputedRadiographyImageStorage | 1.2.840.10008.5.1.4.1.1.1 | sop |
DigitalXRayImageStorageForPresentation | 1.2.840.10008.5.1.4.1.1.1.1 | | sop |
DigitalXRayImageStorageForProcessing | 1.2.840.10008.5.1.4.1.1.1.1.1 | | sop |
DigitalMammographyXRayImageStorageForPresentation | 1.2.840.10008.5.1.4.1.1.1.2 | | sop |
DigitalMammographyXRayImageStorageForProcessing | 1.2.840.10008.5.1.4.1.1.1.2.1 | | sop |
DigitalIntraOralXRayImageStorageForPresentation | 1.2.840.10008.5.1.4.1.1.1.3 | | sop |
DigitalIntraOralXRayImageStorageForProcessing | 1.2.840.10008.5.1.4.1.1.1.3.1 | | sop |
CTImageStorage | 1.2.840.10008.5.1.4.1.1.2 | sop | |
EnhancedCTImageStorage | 1.2.840.10008.5.1.4.1.1.2.1 | | sop |
RETIRED_UltrasoundMultiframeImageStorage | 1.2.840.10008.5.1.4.1.1.3 | sop | |
UltrasoundMultiframeImageStorage | 1.2.840.10008.5.1.4.1.1.3.1 | | sop |
MRImageStorage | 1.2.840.10008.5.1.4.1.1.4 | sop | |
EnhancedMRImageStorage | 1.2.840.10008.5.1.4.1.1.4.1 | | sop |
MRSpectroscopyStorage | 1.2.840.10008.5.1.4.1.1.4.2 | | sop |
EnhancedMRColorImageStorage | 1.2.840.10008.5.1.4.1.1.4.3 | | sop |
RETIRED_NuclearMedicineImageStorage | 1.2.840.10008.5.1.4.1.1.5 | sop | |
RETIRED_UltrasoundImageStorage | 1.2.840.10008.5.1.4.1.1.6 | sop | |
UltrasoundImageStorage | 1.2.840.10008.5.1.4.1.1.6.1 | | sop |
EnhancedUSVolumeStorage | 1.2.840.10008.5.1.4.1.1.6.2 | | sop |
SecondaryCaptureImageStorage | 1.2.840.10008.5.1.4.1.1.7 | sop | |
MultiframeSingleBitSecondaryCaptureImageStorage | 1.2.840.10008.5.1.4.1.1.7.1 | | sop |
MultiframeGrayscaleByteSecondaryCaptureImageStorage | 1.2.840.10008.5.1.4.1.1.7.2 | | sop |
MultiframeGrayscaleWordSecondaryCaptureImageStorage | 1.2.840.10008.5.1.4.1.1.7.3 | | sop |
MultiframeTrueColorSecondaryCaptureImageStorage | 1.2.840.10008.5.1.4.1.1.7.4 | | sop |
RETIRED_StandaloneOverlayStorage | 1.2.840.10008.5.1.4.1.1.8 | sop | |
RETIRED_StandaloneCurveStorage | 1.2.840.10008.5.1.4.1.1.9 | sop | |
DRAFT_WaveformStorage | 1.2.840.10008.5.1.4.1.1.9.1 | | sop |
TwelveLeadECGWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.1.1 | | sop |
GeneralECGWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.1.2 | | sop |
AmbulatoryECGWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.1.3 | | sop |
HemodynamicWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.2.1 | | sop |
CardiacElectrophysiologyWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.3.1 | | sop |
BasicVoiceAudioWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.4.1 | | sop |
GeneralAudioWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.4.2 | | sop |
ArterialPulseWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.5.1 | | sop |
RespiratoryWaveformStorage | 1.2.840.10008.5.1.4.1.1.9.6.1 | | sop |
RETIRED_StandaloneModalityLUTStorage | 1.2.840.10008.5.1.4.1.1.10 | | sop |
RETIRED_StandaloneVOILUTStorage | 1.2.840.10008.5.1.4.1.1.11 | | sop |
GrayscaleSoftcopyPresentationStateStorage | 1.2.840.10008.5.1.4.1.1.11.1 | | sop |
ColorSoftcopyPresentationStateStorage | 1.2.840.10008.5.1.4.1.1.11.2 | | sop |
PseudoColorSoftcopyPresentationStateStorage | 1.2.840.10008.5.1.4.1.1.11.3 | | sop |
BlendingSoftcopyPresentationStateStorage | 1.2.840.10008.5.1.4.1.1.11.4 | | sop |
XAXRFGrayscaleSoftcopyPresentationStateStorage | 1.2.840.10008.5.1.4.1.1.11.5 | | sop |
RETIRED_XASinglePlaneStorage | 1.2.840.10008.5.1.4.1.1.12 | sop | |
XRayAngiographicImageStorage | 1.2.840.10008.5.1.4.1.1.12.1 | | sop |
EnhancedXAImageStorage | 1.2.840.10008.5.1.4.1.1.12.1.1 | | sop |
XRayRadiofluoroscopicImageStorage | 1.2.840.10008.5.1.4.1.1.12.2 | | sop |
EnhancedXRFImageStorage | 1.2.840.10008.5.1.4.1.1.12.2.1 | | sop |
RETIRED_XRayAngiographicBiPlaneImageStorage | 1.2.840.10008.5.1.4.1.1.12.3 | | sop |
XRay3DAngiographicImageStorage | 1.2.840.10008.5.1.4.1.1.13.1.1 | | sop |
XRay3DCraniofacialImageStorage | 1.2.840.10008.5.1.4.1.1.13.1.2 | | sop |
BreastTomosynthesisImageStorage | 1.2.840.10008.5.1.4.1.1.13.1.3 | | sop |
NuclearMedicineImageStorage | 1.2.840.10008.5.1.4.1.1.20 | | sop |
RawDataStorage | 1.2.840.10008.5.1.4.1.1.66 | | sop |
SpatialRegistrationStorage | 1.2.840.10008.5.1.4.1.1.66.1 | | sop |
SpatialFiducialsStorage | 1.2.840.10008.5.1.4.1.1.66.2 | | sop |
DeformableSpatialRegistrationStorage | 1.2.840.10008.5.1.4.1.1.66.3 | | sop |
SegmentationStorage | 1.2.840.10008.5.1.4.1.1.66.4 | | sop |
SurfaceSegmentationStorage | 1.2.840.10008.5.1.4.1.1.66.5 | | sop |
RealWorldValueMappingStorage | 1.2.840.10008.5.1.4.1.1.67 | | sop |
RETIRED_VLImageStorage | 1.2.840.10008.5.1.4.1.1.77.1 | | sop |
VLEndoscopicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.1 | | sop |
VideoEndoscopicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.1.1 | | sop |
VLMicroscopicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.2 | | sop |
VideoMicroscopicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.2.1 | | sop |
VLSlideCoordinatesMicroscopicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.3 | | sop |
VLPhotographicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.4 | | sop |
VideoPhotographicImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.4.1 | | sop |
OphthalmicPhotography8BitImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.5.1 | | sop |
OphthalmicPhotography16BitImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.5.2 | | sop |
StereometricRelationshipStorage | 1.2.840.10008.5.1.4.1.1.77.1.5.3 | | sop |
OphthalmicTomographyImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.5.4 | | sop |
VLWholeSlideMicroscopyImageStorage | 1.2.840.10008.5.1.4.1.1.77.1.6 | | sop |
RETIRED_VLMultiFrameImageStorage | 1.2.840.10008.5.1.4.1.1.77.2 | | sop |
LensometryMeasurementsStorage | 1.2.840.10008.5.1.4.1.1.78.1 | | sop |
AutorefractionMeasurementsStorage | 1.2.840.10008.5.1.4.1.1.78.2 | | sop |
KeratometryMeasurementsStorage | 1.2.840.10008.5.1.4.1.1.78.3 | | sop |
SubjectiveRefractionMeasurementsStorage | 1.2.840.10008.5.1.4.1.1.78.4 | | sop |
VisualAcuityMeasurementsStorage | 1.2.840.10008.5.1.4.1.1.78.5 | | sop |
SpectaclePrescriptionReportStorage | 1.2.840.10008.5.1.4.1.1.78.6 | | sop |
OphthalmicAxialMeasurementsStorage | 1.2.840.10008.5.1.4.1.1.78.7 | | sop |
IntraocularLensCalculationsStorage | 1.2.840.10008.5.1.4.1.1.78.8 | | sop |
MacularGridThicknessAndVolumeReportStorage | 1.2.840.10008.5.1.4.1.1.79.1 | | sop |
OphthalmicVisualFieldStaticPerimetryMeasurementsSt. | 1.2.840.10008.5.1.4.1.1.80.1 | | sop |
DRAFT_SRTextStorage | 1.2.840.10008.5.1.4.1.1.88.1 | | sop |
DRAFT_SRAudioStorage | 1.2.840.10008.5.1.4.1.1.88.2 | | sop |
DRAFT_SRDetailStorage | 1.2.840.10008.5.1.4.1.1.88.3 | | sop |
DRAFT_SRComprehensiveStorage | 1.2.840.10008.5.1.4.1.1.88.4 | | sop |
BasicTextSRStorage | 1.2.840.10008.5.1.4.1.1.88.11 | | sop |
EnhancedSRStorage | 1.2.840.10008.5.1.4.1.1.88.22 | | sop |
ComprehensiveSRStorage | 1.2.840.10008.5.1.4.1.1.88.33 | | sop |
ProcedureLogStorage | 1.2.840.10008.5.1.4.1.1.88.40 | | sop |
MammographyCADSRStorage | 1.2.840.10008.5.1.4.1.1.88.50 | | sop |
KeyObjectSelectionDocumentStorage | 1.2.840.10008.5.1.4.1.1.88.59 | | sop |
ChestCADSRStorage | 1.2.840.10008.5.1.4.1.1.88.65 | | sop |
XRayRadiationDoseSRStorage | 1.2.840.10008.5.1.4.1.1.88.67 | | sop |
ColonCADSRStorage | 1.2.840.10008.5.1.4.1.1.88.69 | | sop |
ImplantationPlanSRDocumentStorage | 1.2.840.10008.5.1.4.1.1.88.70 | | sop |
EncapsulatedPDFStorage | 1.2.840.10008.5.1.4.1.1.104.1 | | sop |
EncapsulatedCDAStorage | 1.2.840.10008.5.1.4.1.1.104.2 | | sop |
PositronEmissionTomographyImageStorage | 1.2.840.10008.5.1.4.1.1.128 | | sop |
RETIRED_StandalonePETCurveStorage | 1.2.840.10008.5.1.4.1.1.129 | | sop |
EnhancedPETImageStorage | 1.2.840.10008.5.1.4.1.1.130 | | sop |
BasicStructuredDisplayStorage | 1.2.840.10008.5.1.4.1.1.131 | | sop |
RTImageStorage | 1.2.840.10008.5.1.4.1.1.481.1 | | sop |
RTDoseStorage | 1.2.840.10008.5.1.4.1.1.481.2 | | sop |
RTStructureSetStorage | 1.2.840.10008.5.1.4.1.1.481.3 | | sop |
RTBeamsTreatmentRecordStorage | 1.2.840.10008.5.1.4.1.1.481.4 | | sop |
RTPlanStorage | 1.2.840.10008.5.1.4.1.1.481.5 | | sop |
RTBrachyTreatmentRecordStorage | 1.2.840.10008.5.1.4.1.1.481.6 | | sop |
RTTreatmentSummaryRecordStorage | 1.2.840.10008.5.1.4.1.1.481.7 | | sop |
RTIonPlanStorage | 1.2.840.10008.5.1.4.1.1.481.8 | | sop |
RTIonBeamsTreatmentRecordStorage | 1.2.840.10008.5.1.4.1.1.481.9 | | sop |
DRAFT_RTBeamsDeliveryInstructionStorage | 1.2.840.10008.5.1.4.34.1 | sop | |
GenericImplantTemplateStorage | 1.2.840.10008.5.1.4.43.1 | sop | |
ImplantAssemblyTemplateStorage | 1.2.840.10008.5.1.4.44.1 | sop | |
ImplantTemplateGroupStorage | 1.2.840.10008.5.1.4.45.1 | sop | |
#GEMRStorage | 1.2.840.113619.4.2 | sop | |
#GECTStorage | 1.2.840.113619.4.3 | sop | |
GE3DModelObjectStorage | 1.2.840.113619.4.26 | sop | |
GERTPlanStorage | 1.2.840.113619.5.249 | sop | |
GERTPlanStorage2 | 1.2.840.113619.4.5.249 | sop | |
GESaturnTDSObjectStorage | 1.2.840.113619.5.253 | sop | |
Philips3DVolumeStorage | 1.2.46.670589.5.0.1 | sop | |
Philips3DObjectStorage | 1.2.46.670589.5.0.2 | sop | |
PhilipsSurfaceStorage | 1.2.46.670589.5.0.3 | sop |
PhilipsCompositeObjectStorage | 1.2.46.670589.5.0.4 | sop |
PhilipsMRCardioProfileStorage | 1.2.46.670589.5.0.7 | sop |
PhilipsMRCardioImageStorage | 1.2.46.670589.5.0.8 | sop |
PatientRootQuery | 1.2.840.10008.5.1.4.1.2.1.1 | sop |
PatientRootRetrieve | 1.2.840.10008.5.1.4.1.2.1.2 | sop |
PatientRootGet | 1.2.840.10008.5.1.4.1.2.1.3 | sop |
StudyRootQuery | 1.2.840.10008.5.1.4.1.2.2.1 | sop |
StudyRootRetrieve | 1.2.840.10008.5.1.4.1.2.2.2 | sop |
StudyRootGet | 1.2.840.10008.5.1.4.1.2.2.3 | sop |
PatientStudyOnlyQuery | 1.2.840.10008.5.1.4.1.2.3.1 | sop |
PatientStudyOnlyRetrieve | 1.2.840.10008.5.1.4.1.2.3.2 | sop |
PatientStudyOnlyGet | 1.2.840.10008.5.1.4.1.2.3.3 | sop |
FindModalityWorkList | 1.2.840.10008.5.1.4.31 | sop |
PatientRootRetrieveNKI | 1.2.826.0.1.3680043.2.135.1066.5.1.4.1.2.1.2 | sop |
StudyRootRetrieveNKI | 1.2.826.0.1.3680043.2.135.1066.5.1.4.1.2.2.2 | sop |
PatientStudyOnlyRetrieveNKI | 1.2.826.0.1.3680043.2.135.1066.5.1.4.1.2.3.2 | sop |
BasicGrayscalePrintManagementMeta | 1.2.840.10008.5.1.1.9 | sop |
BasicColorPrintManagementMeta | 1.2.840.10008.5.1.1.18 | sop |
BasicFilmSession | 1.2.840.10008.5.1.1.1 | sop |
BasicFilmBox | 1.2.840.10008.5.1.1.2 | sop |
BasicGrayscaleImageBox | 1.2.840.10008.5.1.1.4 | sop |
BasicColorImageBox | 1.2.840.10008.5.1.1.4.1 | sop |
BasicPrinter | 1.2.840.10008.5.1.1.16 | sop |
LittleEndianImplicit | 1.2.840.10008.1.2 | transfer | |
#LittleEndianExplicit | 1.2.840.10008.1.2.1 | transfer | |
#BigEndianExplicit | 1.2.840.10008.1.2.2 | transfer | |
#JPEGBaseLine1 | 1.2.840.10008.1.2.4.50 | transfer | LittleEndianExplicit |
#JPEGExtended2and4 | 1.2.840.10008.1.2.4.51 | transfer | LittleEndianExplicit |
#JPEGExtended3and5 | 1.2.840.10008.1.2.4.52 | transfer | LittleEndianExplicit |
#JPEGSpectralNH6and8 | 1.2.840.10008.1.2.4.53 | transfer | LittleEndianExplicit |
#JPEGSpectralNH7and9 | 1.2.840.10008.1.2.4.54 | transfer | LittleEndianExplicit |
#JPEGFulllNH10and12 | 1.2.840.10008.1.2.4.55 | transfer | LittleEndianExplicit |
#JPEGFulllNH11and13 | 1.2.840.10008.1.2.4.56 | transfer | LittleEndianExplicit |
#JPEGLosslessNH14 | 1.2.840.10008.1.2.4.57 | transfer | LittleEndianExplicit |
#JPEGLosslessNH15 | 1.2.840.10008.1.2.4.58 | transfer | LittleEndianExplicit |
#JPEGExtended16and18 | 1.2.840.10008.1.2.4.59 | transfer | LittleEndianExplicit |
#JPEGExtended17and19 | 1.2.840.10008.1.2.4.60 | transfer | LittleEndianExplicit |
#JPEGSpectral20and22 | 1.2.840.10008.1.2.4.61 | transfer | LittleEndianExplicit |
#JPEGSpectral21and23 | 1.2.840.10008.1.2.4.62 | transfer | LittleEndianExplicit |
#JPEGFull24and26 | 1.2.840.10008.1.2.4.63 | transfer | LittleEndianExplicit |
#JPEGFull25and27 | 1.2.840.10008.1.2.4.64 | transfer | LittleEndianExplicit |
#JPEGLossless28 | 1.2.840.10008.1.2.4.65 | transfer | LittleEndianExplicit |
#JPEGLossless29 | 1.2.840.10008.1.2.4.66 | transfer | LittleEndianExplicit |
#JPEGLossless | 1.2.840.10008.1.2.4.70 | transfer | LittleEndianExplicit |
#JPEGLS_Lossless | 1.2.840.10008.1.2.4.80 | transfer | LittleEndianExplicit |
#JPEGLS_Lossy | 1.2.840.10008.1.2.4.81 | transfer | LittleEndianExplicit |
#RLELossless | 1.2.840.10008.1.2.5 | transfer | LittleEndianExplicit |
#LittleEndianExplicitDeflated | 1.2.840.10008.1.2.1.99 | transfer | LittleEndianExplicit |
#JPEG2000LosslessOnly | 1.2.840.10008.1.2.4.90 | transfer | LittleEndianExplicit |
#JPEG2000 | 1.2.840.10008.1.2.4.91 | transfer | LittleEndianExplicit |
APITOKEN EXCHANGES
ALTSWITCH 交易所
No printer configuration options are provided: the default Windows printer is always used. One must use the default document settings of the default printer to change, e.g., the resolution of the printout or the paper size. From 1.4.19b header footer and background bitmaps can be configured through dicom.ini. There is a lua script in folder extensions that improves level and window use for printouts.
On Linux, an experimental print server is Lua is provided that depends on libraries CD and IM, cdlua, imlua. and cdluaim. Enabled by:
[lua]
association = local canvas, canvas_w, canvas_h printserver = dofile('lua/print.lua')
The printer name must be set it the code.
NUKETOKEN LOGIN
The compression settings for dropped images, incoming images, and archival are configured in
dicom.ini. These define the compression mode of images stored on disk by the server.
Dropped images DroppedFileCompression Disk, Remote host IncomingCompression Disk,
Disk ArchiveCompression Archive disk.
The values for these compression settings may be "un" for uncompressed, "as" for as-is (no change in compression), "n1".."n4" for NKI compression styles, "j1".."j2" for loss-less JPEG compression, "j3".."j6" for lossy JPEG compression, “js” for lossless JPEGLS, “j7” for lossy JPEGLS, "jk" for JPEG2000 lossless, "jl" for JPEG2000 lossy, "nj" for NKI or JPEG compression (chooses highest NKI, but leaves JPEG as is), and k1, k2, k4 and.k8 for downsizing to 1024..128 pixels.
The original compression type of incoming images (used with "as") is defined by the remote host, which can choose one of the transfer syntaxes defined in dgatesop.lst.
Since version 1.4.7, if the called AE title in a C-STORE looks like SERVER~xx, then xx will override IncomingCompression (e.g., images sent to a conquest server addressed from the remote host as ‘CONQUESTSRV1~k4’ will be downsized by the server to 256 pixels prior to storage). Note, however, that the total AE may not exceed 16 characters. So this option works correctly only if the base name of the server (CONQUESTSRV1 in the example) has 13 characters or less.
The compression of forwarded images can be set through dicom.ini as well. Disk ExportConverterN Remote host.
The type of compression setting is passed using a command "forward compressed as xx to", where xx is one of the compression types defined for DroppedFileCompression. If the command "forward to" is used instead, the compression type defined in acrnema.map is used. If the remote host does not accept the offered compression, images will automatically be sent with simpler compression or uncompressed. Such negotiation is not implemented for NKI compression.
Images may also be sent as result of a move request to a remote host using different compressions. This option is configured per host in acrnema.map.
Disk Setting in acrnema.map Remote host.
The values for these compression settings may be "un" for uncompressed, "n1".."n4" for NKI compression styles, "j1".."j2" for loss-less JPEG compression, "j3".."j6" for lossy JPEG compression, "jk" for JPEG2000 lossless, "jl" for JPEG2000 lossy, js for lossless JpegLs, j7 for lossy JpegLS, and "k1".."k8" for downsizing the image, 'ul' for littleendianexplicit, 'ub' for bigendianexplicit, and 'ue' for both.
Options "as" and "nj" and “uj” are not correctly implemented for outgoing connections due to the complexity of the transfer syntax negotiation involved. These options may therefore only be
used for NKI clients or the Conquest DICOM server.
If the remote host does not accept the offered JPEG compression, images will automatically be sent with a simpler compression or uncompressed. Such negotiation is not implemented for NKI compression and "k" downsize compression.
Since version 1.4.7, if the called AE title in the C-MOVE looks like SERVER~xx, then xx will override the compression setting in acrnema.map. (E.g., images sent by a conquest server addressed from the remote host as ‘CONQUESTSRV1~k4’ will be downsized by the server to 256 pixels prior to sending).
This allows any host/viewer to receive downsized images on request. Note, however, that the total AE length may not exceed 16 characters. So this option works correctly only if the base name of the server (CONQUESTSRV1 in the example) has 13 characters or less.
For outgoing associations boty uncompressed and compressed syntaxes are proposed. To only propose compressed syntaxes use e.g. “j2!” in acrnema.map.
GOLD BACKED CRYPTO
When dropping a HL7 file onto the server, it initiates the command ‘dgate –loadhl7:file’. This will read the hl7 file and populate the modality worklist database. A sample HL7 file (sample.hl7) is provided for testing. For translating the hl7 data into the DICOM worklist, an extra column has been added to the worklist database definition. Typically this column can contain:
--- No import of values from hl7
*AN Generate a unique 16 character accession number
*UI Generate a unique 64 character UID
SEQ.N Read HL7 sequence seq field N
SEQ.N.M Read HL7 sequence seq field N, subfield M SEQ.N.DATE Read HL7 sequence seq field N, date part SEQ.N.TIME Read HL7 sequence seq field N, time part
Hospitals wanting to use HL7 import should edit this table such that the correct HL7 items are filled in into the worklist database.
When changing the translation part of the worklist database definition, the server must only be restarted to use the adaptations (enable debug log to view the hl7 translation progress). When the database layout of the modality worklist is changed, one should clear the database through the maintenance page and its contents are lost.
Note that database fields marked with ‘DT_STARTSEQUENCE’ and ‘DT_ENDSEQUENCE’ are not used by the program and are descriptive only. The modality worklist query will mimic the organization of the query in sequences in its reply so the sequence organization needs not be specified.
Since version 1.4.16, the accessionnumber is no longer the primary index of the worklist database after it is cleared, allowing more flexibility in filling and using this database.
TUTTI FRUTTI 交易所
Lua scripting provides a wealth of possibilities to configure and extends the functionality of the Conquest DICOM server. Yet programming such scripts may be daunting. By integration of the beautiful ZeroBraneStudio IDE, experimenting with scripting or writing scripts for maintenance tasks becomes much easier. To enable this feature, first download ZeroBraneStudio from (http://studio.zerobrane.com) or get the latest repository as ZIP file from Github (https://github.com/pkulchenko/ZeroBraneStudio). Unzip the zip file, the executable zbstudio.exe is ready to go. Start it. Then to integrate Conquest dicom server, open from the c:\dicomserver folder the file ZeroBraneStudio\install.lua in ZeroBraneStudio.
Open ZeroBraneStudio and look at the Local console tab Load this install.lua file with File – Open or drag and drop Click the [install] link to do finish installation.
After this - ZeroBrane Studio will reopen ready to run demo scripts and develop for Conquest Dicom Server. As installed, Zerobrane Studio communicates with a running Conquest Dicom Server and offers code
completion and full debugging facilities. Try for instance to enter a small query script:
a=DicomObject:new() a.QueryRetrieveLevel='STUDY' a.StudyInstanceUID='' a.PatientID='' a.StudyDate='19980101-19990101'
b=dicomquery('CONQUESTSRV1', 'STUDY', a) for i=0, #b-1 do
print(b[i].PatientID, b[i].StudyInstanceUID, '\n') end
Run it with F5-F5. For a default server, this will print in the 'Output' tab:
Program completed in 0.17 seconds (pid: 5948). Debugging session started in 'C:\dicomserver\'.
"0009703828" "1.3.46.670589.5.2.10.2156913941.892665384.993397" "\n"
Debugging session completed (traced 0 instructions).
Note that this script is run by the Conquest DICOM server, which has to be up for this script to work. If you change project - Lua interpreter to Conquest DICOM Utility, a new instance of dgate.exe will be run to run the script, and the output will be:
Program starting as '"C:\dicomserver\dgate.exe" --dolua:"xpcall(function() io.stdout:setvbuf('no');
require('mobdebug').yield=function() if iup then iup.LoopStep() end end; require('mobdebug').loop('CF-J10',8172) ;
package.loaded.mobdebug.done() ; package.loaded.mobdebug=nil ;
collectgarbage('collect') end, function(err) print(debug.traceback(err)) end)"'.
Program 'dgate.exe' started in 'C:\dicomserver' (pid: 5212). Debugging session started in 'C:\dicomserver\'.
0009703828 1.3.46.670589.5.2.10.2156913941.892665384.993397
Debugging session completed (traced 0 instructions). Program completed in 2.49 seconds (pid: 5212).
In the Lua script, all normal Lua functionality is present (the examples delivered with ZeroBraneStudio are a good starting point to learn Lua), and the following variables and classes are defined:
Filename Name of dropped file (if any)
command_line command from importconverter e.g., “process series by t.lua command”
returnfile If set from lua server command; file contents are returned to client
Global Server status (counters) and configuration
e.g.: Global.StartTime Global.BaseDir
Association Information of current connection
Association.Calling Association.Called Association.Thread Association.ConnectedIP
Command Received command by server Command.AffectedSOPClassUID Command.AffectedSOPInstanceUID
Command.CommandField Command.MessageID Command.MessageIDBeingRespondedTo Command.DataSetType Command.MoveDestination Command.TransferSyntaxUID
Command.MoveOriginatorApplicationEntityTitle Command.MoveOriginatorMessageID Command:Write(filename) write dicom object
Command:Dump(filename) write dicom object header as text file
It is possible to add modify the Command and/or add private Conquest tags to modify e.g. move behavior. See e.g. ConquestMaxSlices, ConquestScript, ConquestSplit
Data Data object of current DICOM command
(DicomObject)
DicomObject DICOM Object (e.g., image) DicomObject:new() returns empty DicomObject DicomObject:new(json_string) convert json to DicomObject DicomObject:newarray() returns empty DicomArray
DicomObject:free () frees DicomObject or DicomArray
Data.Storagestring e.g. MAG0 for importconverters
DicomObject:Script(code) run conquest style script DicomObject:GetPixel(x, y, fr) get pixel returns 1..N values DicomObject:SetPixel(x, y, fr, values) set pixel DicomObject:GetRow(y, frame) get row returning array DicomObject:SetRow(y, frame, table) set row of pixels DicomObject:GetColumn(x, fr) get column returns array DicomObject:SetColumn(x, fr, table) set column of pixels DicomObject:GetImage(frame) get image as binary string DicomObject:SetImage(frame, string) set 2D image in dicom object DicomObject:SetImage(frame,string,scale) set 2D float image object DicomObject:Read(filename: string) read dicom object
Note: do not use Data:Read DicomObject:Write(filename: string) write dicom object
DicomObject:Dump(filename: string) write header as text file DicomObject:GetVR(grp, elmnt, asstring) get vr as byte sequence returns
DicomArray/table/string DicomObject:SetVR(grp, elmnt, value) set vr from DicomArray/table of
bytes/binary/json string
DicomObject:AddImage() Enter image into DICOM server
DicomObject:Copy() Returns copy of object DicomObject:Copy(json_string) Combine object with json DicomObject:Compress(string) Returns compressed copy object DicomObject:Serialize() Returns object in Lua syntax
DicomObject:Serialize(true) Returns object in JSON syntax DicomObject:Serialize(true,true) Returns in JSON with pixeldata DicomObject:Serialize(true,true,true) Returns in DicomWEB JSON syntax Data.PatientID Any VR is accessible
DicomArray Dicom sequence = array of DICOM objects DicomObject:newarray() create
DicomArray:free() free
DicomArray:Add(dicomobject) add dicomobject to array
DicomArray:Delete(0) Delete numbered entry
DicomArray:Serialize() Returns object in Lua syntax
DicomArray:Serialize(true) Returns object in JSON syntax
DicomArray[0] elements 0..#-1 are DicomObject
Utility functions
newdicomobject() returns DicomObject
deletedicomobject(DicomObject) free DicomObject
newdicomarray() returns DicomArray
print(...) print arguments to console
debuglog(...) print if debug logging enabled
gpps(section, key, default) Reads DICOM.INI
crc(string) Calculates crc of string
tickcount() Return time in ms
dictionary(group, element) return dictionary name
dictionary(name) returns dict group, element get_sqldef(database, field: integer) Reads DICOM.SQL returns
(group, element, FieldName, Length, SQLType, DicomType system(program: string) run program in the background listoldestpatients(max:integer, device:string, sort:string)
Find oldest patients e.g. to move/delete, sort on database field; returns list of patID's
destroy() Special for server events
reject() See e.g. script('destroy')
retry()
genuid() Returns new UID
tempfile(extension: string) Returns temp file name
get_amap(entry: int) Reads item from acrnema.map returns AE, IP, port, compression
dbquery(database, fields, query) Executes SQL query on database returns table of records from 1 with table of fields from 1 return nil if query fails; {} if query result empty
dicomquery(AE, level, query: DicomObject) returns sequence counting from 0 dicomquery2(AE, level, query: DicomObject) (old) returns sequence from 0 dicommove(AE, dest, query: DicomObject, patroot:number, extra:DicomObject)
move data from DICOM server to DICOM server, extra tags can change the outgoing images
dicomget(AE:string, level:string, query) get object array from server dicomread(query) same from local server dicomdelete( query: DicomObject) delete data from DICOM server dicomstore(ae:string; DicomObject/DicomArray) send data to any DICOM server dicomecho(ae:string; extra:DicomObject) echo server, reads raw response dicomprint(Object/Array,AE,format,callback) print object(array) to server heapinfo() returns string allocations
sql(statement: string) execute SQL statement
changeuid(olduid[, proposeduid] Consistently modify UID, returns mapped UID changeuid('') clears UID cache
changeuidback(newuid: string) For a modified UID, returns original if exists, returns (string or nil)\
addimage(image: userdata) Enter image into server
sleep(N: integer) Sleep for N ms
mkdir(path: string) Make directory (recursive) ConvertBinaryData(format:string, data:string|table)
convert table to binary string or vice-versa; format e.g. f8; 1000*f8; 1000*u4, 100*s100
HTML(text: string, …) web server only: output HTML
CGI(key, default) web server only: read url entry
CGI() web server only: rd post buffer
servercommand(command: string, mode:string) Sends conquest server command,
e.g. 'display_status:' or 'get_param:MyACRNema' (string) mode can 'cgibinary', 'cgihtml' or <filename to upload and
>filename to download, 'binary' without procesing.
The following ways allow returning data fom server to script:
lua:return expression – limited to 512 characters lua:Data:SetVR(0x9999,0x0401,longstring)
lua:local s=tempfile('txt') local f=io.open(s, "wb") f:write(r) returnfile=s f:close();
servercommand('luastart:statement') starts a new thread
These functions work on variable Data
script(script_code: string) Sends conquest style script to server, e.g. 'forward to AE'. Special functions are: script('retry') for RejectedImageWorkListConverter0 and RejectedImageConverter0; will re-attempt to store the object after the script is done, script('defer') for ExportConverter: will cause later retry, and script('destroy') for query/store or move: will cancel operation; script('destroy2') will do the same silently
getpixel(x: int, y: int, frame: int) returns pixel values setpixel(x: int, y: int, frame: int, pixel: int, …) set pixel values getrow(y: int, frame: int) get row of pixels as
arraysetrow(y: int, frame: int, table) set row of pixels getcolumn(x: int, frame: int) get column of pixels as array setcolumn(x: int, frame: int, a: table) set column of pixels getimage(frame: int) get image as binary string
setimage(frame: int, a: string) set entire image setimage(frame: int, a: string, scale:float) set short image from floats getvr(group, element, asstring) get VR as DicomArray/byte
array/binary string from dicom object returns (DicomArray/table/string of VR values)
setvr(group, element, a) set VR sequence or binary
readdicom(filename) read dicom object
writedicom(filename) write dicom object
writeheader(filename) write header as text file
Note that calls to the Conquest script language, e.g. script() and Data:Script() can do forward calls e.g.
Data:Script('forward to AE').
These calls will share a single channel (19.0) so it is best to restrict your script with just one of these calls, so that the forward code can optimise transfers.





















